4Y95
 
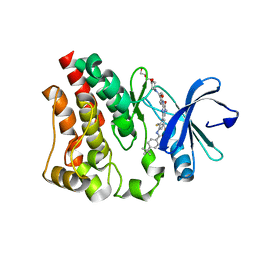 | | Crystal structure of the kinase domain of Bruton's tyrosine kinase with mutations in the activation loop | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, BETA-MERCAPTOETHANOL, ... | | Authors: | Wang, Q, Rosen, C.E, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
4IRV
 
 | |
8T8K
 
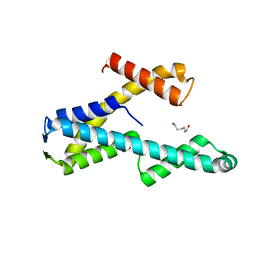 | |
8T8L
 
 | |
2N4L
 
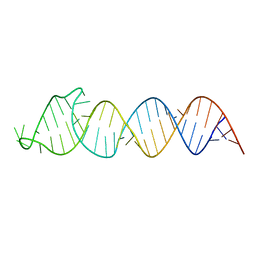 | | Solution Structure of the HIV-1 Intron Splicing Silencer and its Interactions with the UP1 Domain of hnRNP A1 | | Descriptor: | RNA (53-MER) | | Authors: | Tolbert, B.S, Jain, N, Morgan, C.E, Rife, B.D, Salemi, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Intron Splicing Silencer and Its Interactions with the UP1 Domain of Heterogeneous Nuclear Ribonucleoprotein (hnRNP) A1.
J.Biol.Chem., 291, 2016
|
|
4LOS
 
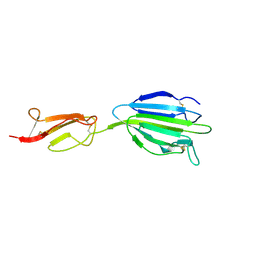 | | C1s CUB2-CCP1 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E, Gingras, A.R. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOT
 
 | | C1s CUB2-CCP1-CCP2 | | Descriptor: | Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4YAY
 
 | | XFEL structure of human Angiotensin Receptor | | Descriptor: | 5,7-diethyl-1-{[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl}-3,4-dihydro-1,6-naphthyridin-2(1H)-one, Soluble cytochrome b562,Type-1 angiotensin II receptor | | Authors: | Zhang, H, Unal, H, Gati, C, Han, G.W, Zatsepin, N.A, James, D, Wang, D, Nelson, G, Weierstall, U, Messerschmidt, M, Williams, G.J, Boutet, S, Yefanov, O.M, White, T.A, Liu, W, Ishchenko, A, Tirupula, K.C, Desnoyer, R, Sawaya, M.C, Xu, Q, Coe, J, Cornrad, C.E, Fromme, P, Stevens, R.C, Katritch, V, Karnik, S.S, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Angiotensin receptor revealed by serial femtosecond crystallography.
Cell, 161, 2015
|
|
4JS5
 
 | |
5G3Q
 
 | | Crystal structure of a hypothetical domain in WNK1 | | Descriptor: | WNK1 | | Authors: | Pinkas, D.M, Bufton, J.C, Sanvitale, C.E, Bartual, S.G, Adamson, R.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | Crystal Structure of a Hypothetical Domain in Wnk1
To be Published
|
|
4X88
 
 | | E178D Selectivity filter mutant of NavMS voltage-gated pore | | Descriptor: | HEGA-10, Ion transport protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
5FFX
 
 | |
5F92
 
 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fumarate hydratase class II, ... | | Authors: | Kasbekar, M, Fischer, G, Mott, B.T, Yasgar, A, Hyvonen, M, Boshoff, H.I, Abell, C, Barry, C.E, Thomas, C.J. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Selective small molecule inhibitor of the Mycobacterium tuberculosis fumarate hydratase reveals an allosteric regulatory site.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4JJM
 
 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YX5
 
 | | SpaO(SPOA1,2) | | Descriptor: | CHLORIDE ION, Surface presentation of antigens protein SpaO | | Authors: | Notti, R.Q, Stebbins, C.E. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9001 Å) | | Cite: | A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Nat Commun, 6, 2015
|
|
4YFK
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
8RTZ
 
 | | The structure of E. coli penicillin binding protein 3 (PBP3) in complex with a bicyclic peptide inhibitor | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Bicyclic peptide inhibitor, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Rowland, C.E, Dods, R, Lewis, N, Stanway, S.J, Bellini, D, Beswick, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and chemical optimisation of a Potent, Bi-cyclic (Bicycle) Antimicrobial Inhibitor of Escherichia coli PBP3
To Be Published
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
5FFZ
 
 | |
4MB4
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JS4
 
 | |
5FB2
 
 | |
4MB5
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8T4O
 
 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF347 inhibitor with no glutamate | | Descriptor: | 4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoic acid, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-06-09 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
