3PQK
 
 | |
8FWN
 
 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|
8FWO
 
 | | Crystal structure of SARS-CoV-2 papain-like protease | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease
To Be Published
|
|
4JJM
 
 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
7UXX
 
 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7UXZ
 
 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
3PQJ
 
 | |
2PXG
 
 | | NMR Solution Structure of OmlA | | Descriptor: | Outer membrane protein | | Authors: | Vanini, M.M.T, Pertinhez, T.A, Sforca, M.L, Spisni, A, Benedetti, C.E. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the outer membrane lipoprotein OmlA from Xanthomonas axonopodis pv. citri reveals a protein fold implicated in protein-protein interaction.
Proteins, 71, 2008
|
|
5U50
 
 | |
5U4Z
 
 | | Crystal structure of citrus MAF1 in space group P 31 2 1 | | Descriptor: | Repressor of RNA polymerase III transcription, SULFATE ION | | Authors: | Soprano, A.S, Giuseppe, P.O, Nascimento, A.F.Z, Benedetti, C.E, Murakami, M.T. | | Deposit date: | 2016-12-06 | | Release date: | 2017-07-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of citrus MAF1 in space group P 31 2 1
To Be Published
|
|
2M70
 
 | |
2NUH
 
 | |
6WQ6
 
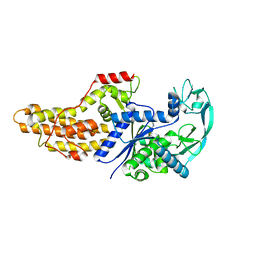 | |
6WQS
 
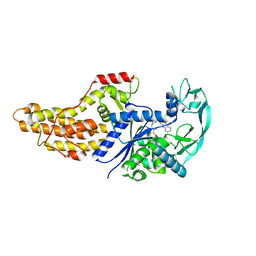 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP8839 | | Descriptor: | 2-{[3-({[4-bromo-5-(1-fluoroethenyl)-3-methylthiophen-2-yl]methyl}amino)propyl]amino}quinolin-4(1H)-one, Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
6WQT
 
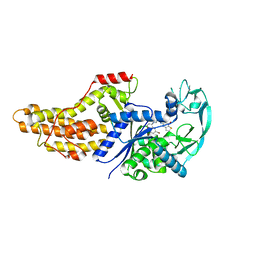 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP3123 | | Descriptor: | 5-[(3-{[(4R)-6,8-dibromo-3,4-dihydro-2H-1-benzopyran-4-yl]amino}propyl)amino]thieno[3,2-b]pyridin-7(6H)-one, Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
6WQI
 
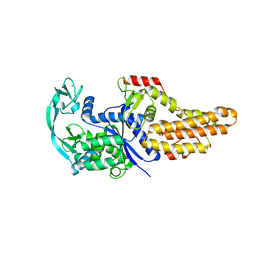 | | Xanthomonas citri Methionyl-tRNA synthetase (apo) | | Descriptor: | Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
2KQA
 
 | | The solution structure of the fungal elicitor Cerato-Platanin | | Descriptor: | Cerato-platanin | | Authors: | Oliveira, A.L, Gallo, M, Pazzagli, L, Cappugi, G, Scala, A, Cicero, D.O, Pantera, B, Spisni, A, Benedetti, C.E, Pertinhez, T.A. | | Deposit date: | 2009-11-03 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the fungal elicitor Cerato-Platanin
To be Published
|
|
2KQ5
 
 | |
