8V9H
 
 | | GES-5-NA-1-157 complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
2O6E
 
 | | Structure of native rTp34 from Treponema pallidum from zinc-soaked crystals | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2OIV
 
 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
1G1D
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2-FLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2-FLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-11 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Contribution of Fluorine to Protein-Ligand Affinity in the Binding of Fluoroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
2NYI
 
 | | Crystal Structure of an Unknown Protein from Galdieria sulphuraria | | Descriptor: | unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tandem ACT domain-containing protein ACTP from Galdieria sulphuraria.
Proteins, 80, 2012
|
|
1G4O
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-PHENYLMETHYLBENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-BENZYL-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
2NXL
 
 | |
1D5L
 
 | | CRYSTAL STRUCTURE OF CYANIDE-BOUND HUMAN MYELOPEROXIDASE ISOFORM C AT PH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Fiedler, T.J, Davey, C.A, Fenna, R.E. | | Deposit date: | 1999-10-07 | | Release date: | 2001-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human myeloperoxidase: structure of a cyanide complex and its interaction with bromide and thiocyanate substrates at 1.9 A resolution.
Biochemistry, 40, 2001
|
|
1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
2OIX
 
 | | Xanthomonas XopD C470A Mutant | | Descriptor: | Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
3BIU
 
 | | Human thrombin-in complex with UB-THR10 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentylamino)ethanoyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, ... | | Authors: | Gerlach, C, Smolinski, M, Steuber, H, Sotriffer, C.A, Heine, A, Hangauer, D.G, Klebe, G. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Inhibition Profile of a Cyclopentyl and a Cyclohexyl Derivative towards Thrombin: The Same but for Different Reasons
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
1G45
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2-FLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2-FLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-26 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1G4J
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2,3,4,5,6-PENTAFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1FZH
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II PRESSURIZED WITH XENON GAS | | Descriptor: | CALCIUM ION, FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1G46
 
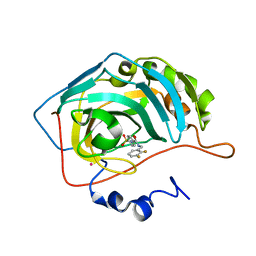 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3-DIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2,3-DIFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-26 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
2O6C
 
 | | Structure of selenomethionyl rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
3B6F
 
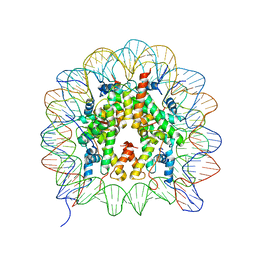 | |
1CJG
 
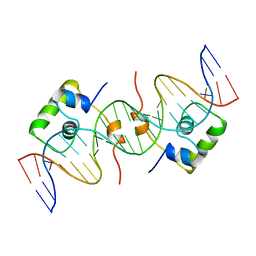 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
2YN8
 
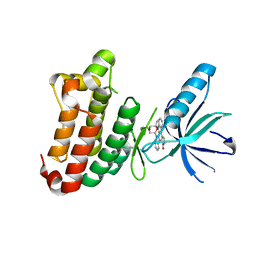 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE | | Authors: | Read, J, Brassington, C.A, Overmann, R. | | Deposit date: | 2012-10-13 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Stability and Solubility Engineering of the Ephb4 Tyrosine Kinase Catalytic Domain Using a Rationally Designed Synthetic Library.
Protein Eng.Des.Sel., 26, 2013
|
|
1DQL
 
 | | CRYSTAL STRUCTURE OF AN UNLIGANDED (NATIVE) FV FROM A HUMAN IGM ANTI-PEPTIDE ANTIBODY | | Descriptor: | IGM MEZ IMMUNOGLOBULIN | | Authors: | Ramsland, P.A, Shan, L, Moomaw, C.R, Slaughter, C.A, Guddat, L.W, Edmundson, A.B. | | Deposit date: | 2000-01-04 | | Release date: | 2000-10-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An unusual human IgM antibody with a protruding HCDR3 and high avidity for its peptide ligands.
Mol.Immunol., 37, 2000
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
2PSU
 
 | | Crystal Structure of wild type HIV-1 protease in complex with CARB-AD37 | | Descriptor: | ACETATE ION, N-[(1S,2R)-1-BENZYL-3-{(CYCLOPROPYLMETHYL)[(3-METHOXYPHENYL)SULFONYL]AMINO}-2-HYDROXYPROPYL]-N'-METHYLSUCCINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Mutation-resistant HIV Protease Inhibitors with the Substrate Envelope Hypothesis.
Chem.Biol.Drug Des., 69, 2007
|
|
1DNH
 
 | | THE MOLECULAR STRUCTURE OF THE COMPLEX OF HOECHST 33258 AND THE DNA DODECAMER D(CGCGAATTCGCG) | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Teng, M.-K, Usman, N, Frederick, C.A, Wang, A.H.-J. | | Deposit date: | 1988-02-16 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG).
Nucleic Acids Res., 16, 1988
|
|
1GFW
 
 | |
1G3Z
 
 | | CARBONIC ANHYDRASE II (F131V) | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, ZINC ION | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-25 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Contribution of Fluorine to Protein-Ligand Affinity in the Binding of Fluoroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
