1LKV
 
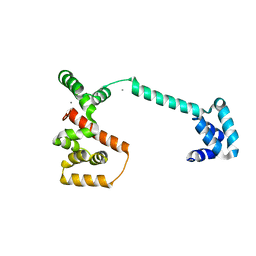 | |
2HUO
 
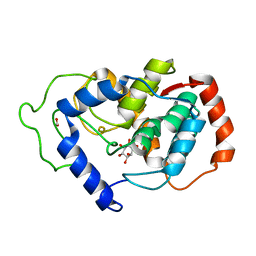 | | Crystal structure of mouse myo-inositol oxygenase in complex with substrate | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | Authors: | Brown, P.M, Caradoc-Davies, T.T, Dickson, J.M.J, Cooper, G.J.S, Loomes, K.M, Baker, E.N. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1HKY
 
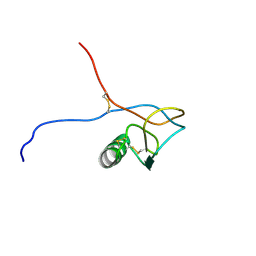 | | Solution structure of a PAN module from Eimeria tenella | | Descriptor: | MICRONEME PROTEIN 5 PRECURSOR | | Authors: | Brown, P.J, Mulvey, D, Potts, J.R, Tomley, F.M, Campbell, I.D. | | Deposit date: | 2002-10-03 | | Release date: | 2002-10-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Pan Module from the Apicomplexan Parasite Eimeria Tenella
J.Struct.Funct.Genom., 4, 2003
|
|
1HLO
 
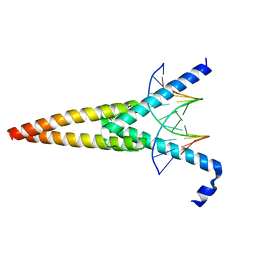 | | THE CRYSTAL STRUCTURE OF AN INTACT HUMAN MAX-DNA COMPLEX: NEW INSIGHTS INTO MECHANISMS OF TRANSCRIPTIONAL CONTROL | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*CP*CP*AP*CP*GP*TP*GP*GP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR MAX) | | Authors: | Brownlie, P, Ceska, T.A, Lamers, M, Romier, C, Theo, H, Suck, D. | | Deposit date: | 1997-09-10 | | Release date: | 1997-10-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control.
Structure, 5, 1997
|
|
7M40
 
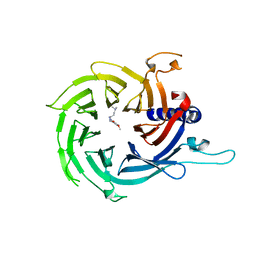 | | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4) | | Descriptor: | Histone-binding protein RBBP4, N~3~-{4-[3-(dimethylamino)pyrrolidin-1-yl]-6,7-dimethoxyquinazolin-2-yl}-N~1~,N~1~-dimethylpropane-1,3-diamine | | Authors: | Perveen, S, Dong, A, Tempel, W, Zepeda-Velazquez, C, Abbey, M, McLeod, D, Marcellus, R, Mohammed, M, Ensan, D, Panagopoulos, D, Trush, V, Gibson, E, Brown, P.J, Arrowsmith, C.H, Schapira, M, Al-awar, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4)
To Be Published
|
|
3K5K
 
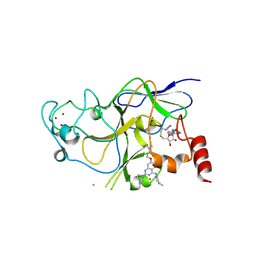 | | Discovery of a 2,4-Diamino-7-aminoalkoxy-quinazoline as a Potent Inhibitor of Histone Lysine Methyltransferase, G9a | | Descriptor: | 7-[3-(dimethylamino)propoxy]-6-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinazolin-4-amine, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Dong, A, Wasney, G.A, Liu, F, Chen, X, Allali-Hassani, A, Senisterra, G, Chau, I, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Frye, S.V, Bochkarev, A, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a.
J.Med.Chem., 52, 2009
|
|
5W1Y
 
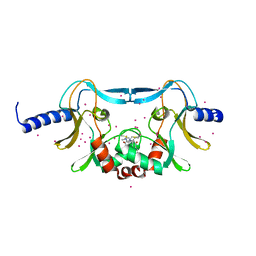 | | SETD8 in complex with a covalent inhibitor | | Descriptor: | 2-(4-methylpiperazin-1-yl)-3-(phenylsulfanyl)naphthalene-1,4-dione, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Yu, W, Li, Y, Blum, G, Luo, M, Pittella-Silva, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETD8 in complex with a covalent inhibitor
to be published
|
|
4MI0
 
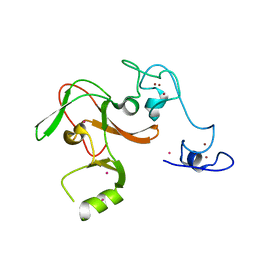 | | Human Enhancer of Zeste (Drosophila) Homolog 2(EZH2) | | Descriptor: | Histone-lysine N-methyltransferase EZH2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations.
Plos One, 8, 2013
|
|
6UE6
 
 | | PWWP1 domain of NSD2 in complex with MR837 | | Descriptor: | 4-cyano-N-cyclopropyl-N-[(thiophen-2-yl)methyl]benzamide, Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, De Freitas, R.F, Schapira, M, Brown, P.J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the PWWP Domain of NSD2.
J.Med.Chem., 64, 2021
|
|
7UFV
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
7RBQ
 
 | | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9, ... | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor
To Be Published
|
|
8G38
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
7U9I
 
 | | Co-crystal structure of human CARM1 in complex with MT556 inhibitor | | Descriptor: | 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Perveen, S, Dong, A, Hutchinson, A, Seitova, A, Gibson, E, Hajian, T, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human CARM1 in complex with MT556 inhibitor
To Be Published
|
|
8EOM
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973 | | Descriptor: | 4-(4-methylpiperazine-1-sulfonyl)benzamide, SULFATE ION, TP53-binding protein 1, ... | | Authors: | The, J, Hong, Z, Headey, S, Gunzburg, M, Doak, B, James, L.I, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973
to be published
|
|
8F0W
 
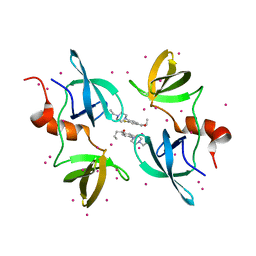 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
8T55
 
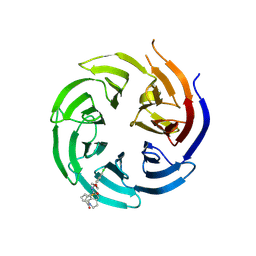 | | Co-crystal structure of the WD-repeat domain of human WDR91 in complex with MR46654 | | Descriptor: | 1,2-ETHANEDIOL, N-[3-(4-chlorophenyl)oxetan-3-yl]-1-propanoyl-1,2,3,4-tetrahydroquinoline-5-carboxamide, WD repeat-containing protein 91 | | Authors: | Ahmad, H, Zeng, H, Dong, A, Li, Y, Yen, H, Seitova, A, Xu, J, Feng, J.W, Brown, P.J, Santhakumar, V, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
8SHJ
 
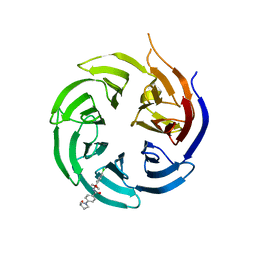 | | Crystal structure of the WD-repeat domain of human WDR91 in complex with MR45279 | | Descriptor: | N-[3-(4-chlorophenyl)oxetan-3-yl]-4-[(3S)-3-hydroxypyrrolidin-1-yl]benzamide, WD repeat-containing protein 91 | | Authors: | Ahmad, H, Zeng, H, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Xu, J, Feng, J.W, Brown, P.J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
8T5I
 
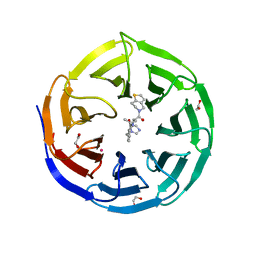 | | Crystal structure of human WDR5 in complex with MR4397 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(2S)-1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl]-3-[(1H-imidazol-1-yl)methyl]benzamide, ... | | Authors: | Kimani, S, Dong, A, Li, F, Loppnau, P, Ackloo, S, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human WDR5 in complex with MR4397
To be published
|
|
8T68
 
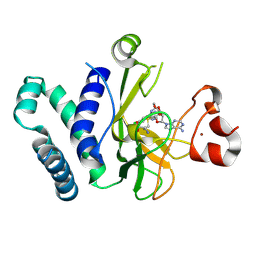 | | Crystal Structure of the SET Domain of Human Histone-Lysine N-Methyltransferase SUV420H1 in complex with RQ3-111 | | Descriptor: | 6,7-dichloro-N-[(3R)-oxolan-3-yl]-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase KMT5B, S-ADENOSYLMETHIONINE, ... | | Authors: | Zeng, H, Dong, A, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-15 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the SET Domain of Human Histone-Lysine N-Methyltransferase SUV420H1 in complex with RQ3-111.
To be published
|
|
8UWP
 
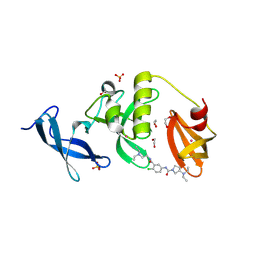 | | Crystal structure of SETDB1 Tudor domain in complex with MR46747 | | Descriptor: | (3S)-N-(4-chloro-3-{[2-(diethylamino)ethyl]carbamoyl}phenyl)-3-(diethylamino)pyrrolidine-1-carboxamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Shrestha, S, Beldar, S, Dong, A, Ackloo, S, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-07 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with MR46747
To be published
|
|
8G5E
 
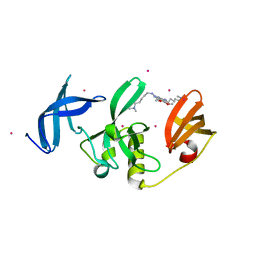 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, N~4~-[6-(dimethylamino)hexyl]-N~2~-[5-(dimethylamino)pentyl]-6,7-dimethoxyquinazoline-2,4-diamine, UNKNOWN ATOM OR ION | | Authors: | Beldar, S, Dong, A, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535
To be published
|
|
