7NLU
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(1H-indol-3-yl)ethan-1-one | | Descriptor: | 1-(1~{H}-indol-3-yl)ethanone, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN7
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with dimethyl 5-hydroxyisophthalate. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with methyl 4-hydroxy-3-iodobenzoate. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION, methyl 3-iodanyl-4-oxidanyl-benzoate | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
1PDA
 
 | | STRUCTURE OF PORPHOBILINOGEN DEAMINASE REVEALS A FLEXIBLE MULTIDOMAIN POLYMERASE WITH A SINGLE CATALYTIC SITE | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, PORPHOBILINOGEN DEAMINASE | | Authors: | Louie, G.V, Brownlie, P.D, Lambert, R, Cooper, J.B, Blundell, T.L, Wood, S.P, Warren, M.J, Woodcock, S.C, Jordan, P.M. | | Deposit date: | 1992-11-17 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site.
Nature, 359, 1992
|
|
7ORZ
 
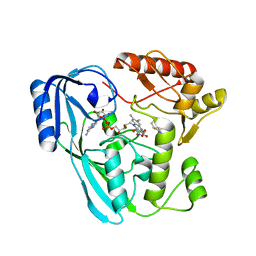 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-06 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OR2
 
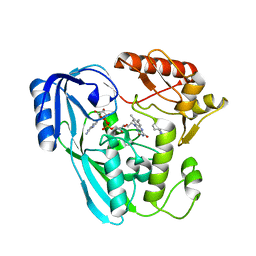 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OSQ
 
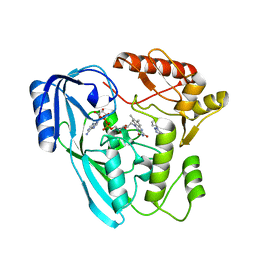 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OTV
 
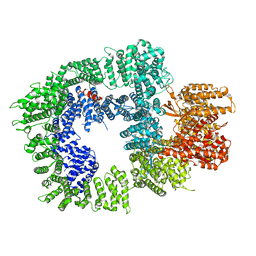 | | DNA-PKcs in complex with wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTW
 
 | | DNA-PKcs in complex with AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTM
 
 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTY
 
 | | DNA-PKcs in complex with M3814 | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTP
 
 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
4MUE
 
 | |
4MUF
 
 | |
4MUN
 
 | |
4MQ6
 
 | |
4MUH
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(2-(5-acetamido-1,3,4-thiadiazol-2-ylsulfonylcarbamoyl)-5-methoxy-1H-indol-1-yl)acetic acid | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Pantothenate synthetase, ... | | Authors: | Silvestre, H.L, Blundell, T.L. | | Deposit date: | 2013-09-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Optimization of Inhibitors of Mycobacterium tuberculosis Pantothenate Synthetase Based on Group Efficiency Analysis.
Chemmedchem, 11, 2016
|
|
4KIU
 
 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49d [5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
1N0W
 
 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
4MUG
 
 | |
4KG0
 
 | | Crystal structure of the drosophila melanogaster neuralized-nhr1 domain | | Descriptor: | MAGNESIUM ION, Protein neuralized | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2013-04-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure, biochemical and biophysical characterisation of NHR1 domain of E3 Ubiquitin ligase neutralized
Advances in Enzyme Research, 1, 2013
|
|
4KM2
 
 | |
1MPP
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
4KM0
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with pyrimethamine | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
1PWA
 
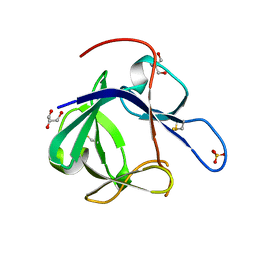 | | Crystal structure of Fibroblast Growth Factor 19 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fibroblast growth factor-19, GLYCEROL, ... | | Authors: | Harmer, N.J, Pellegrini, L, Chirgadze, D, Fernandez-Recio, J, Blundell, T.L. | | Deposit date: | 2003-07-01 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of fibroblast growth factor (FGF) 19 reveals novel features of the FGF family and offers a structural basis for its unusual receptor affinity.
Biochemistry, 43, 2004
|
|
