1MPP
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
3CMS
 
 | | ENGINEERING ENZYME SUB-SITE SPECIFICITY: PREPARATION, KINETIC CHARACTERIZATION AND X-RAY ANALYSIS AT 2.0-ANGSTROMS RESOLUTION OF VAL111PHE SITE-MUTATED CALF CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Shearer, A, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1990-02-26 | | Release date: | 1992-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering enzyme subsite specificity: preparation, kinetic characterization, and X-ray analysis at 2.0-A resolution of Val111Phe site-mutated calf chymosin.
Biochemistry, 29, 1990
|
|
2BGW
 
 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BHN
 
 | | XPF from Aeropyrum pernix | | Descriptor: | XPF ENDONUCLEASE | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
1DMU
 
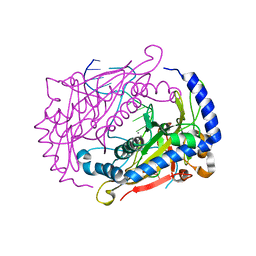 | | Crystal structure of the restriction endonuclease BglI (e.c.3.1.21.4) bound to its dna recognition sequence | | Descriptor: | BETA-MERCAPTOETHANOL, BGLI RESTRICTION ENDONUCLEASE, CALCIUM ION, ... | | Authors: | Newman, M, Lunnen, K, Wilson, G, Greci, J, Schildkraut, I, Phillips, S.E.V. | | Deposit date: | 1999-12-15 | | Release date: | 1999-12-18 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence.
EMBO J., 17, 1998
|
|
4CMS
 
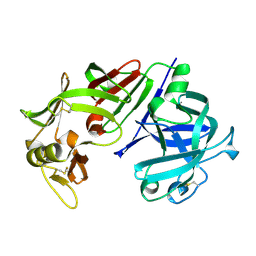 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES IV. STRUCTURE AND REFINEMENT AT 2.2 ANGSTROMS RESOLUTION OF BOVINE CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Khan, G, Tickle, I.J, Blundell, T.L, Safro, M, Andreeva, N, Zdanov, A. | | Deposit date: | 1991-11-01 | | Release date: | 1991-11-07 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray analyses of aspartic proteinases. IV. Structure and refinement at 2.2 A resolution of bovine chymosin.
J.Mol.Biol., 221, 1991
|
|
1BAM
 
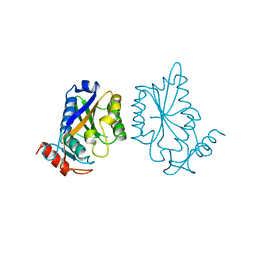 | |
1BHM
 
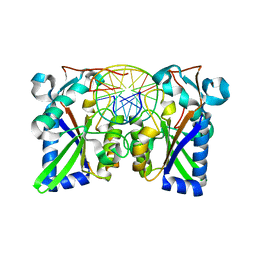 | | RESTRICTION ENDONUCLEASE BAMHI COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*TP*AP*TP*GP*GP*AP*TP*CP*CP*AP*TP*A)-3'), PROTEIN (BAMHI (E.C.3.1.21.4)) | | Authors: | Aggarwal, A.K, Newman, M. | | Deposit date: | 1995-07-12 | | Release date: | 1995-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding.
Science, 269, 1995
|
|
1L8G
 
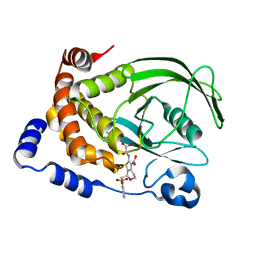 | | Crystal structure of PTP1B complexed with 7-(1,1-Dioxo-1H-benzo[d]isothiazol-3-yloxymethyl)-2-(oxalyl-amino)-4,7-dihydro-5H-thieno[2,3-c]pyran-3-carboxylic acid | | Descriptor: | 7-(1,1-DIOXO-1H-BENZO[D]ISOTHIAZOL-3-YLOXYMETHYL)-2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Iversen, L.F, Andersen, H.S, Moller, K.B, Olsen, O.H, Peters, G.H, Branner, S, Mortensen, S.B, Hansen, T.K, Lau, J, Ge, Y, Holsworth, D.D, Newman, M.J, Moller, N.P.H. | | Deposit date: | 2002-03-20 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steric hindrance as a basis for structure-based design of selective inhibitors of protein-tyrosine phosphatases.
Biochemistry, 40, 2001
|
|
2JXR
 
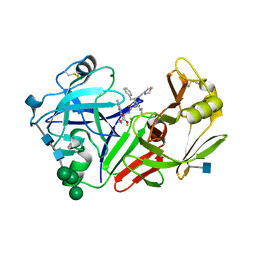 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
