6QRA
 
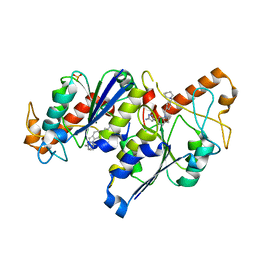 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-[1-[[4-(piperidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazol-5-amine, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
6QR0
 
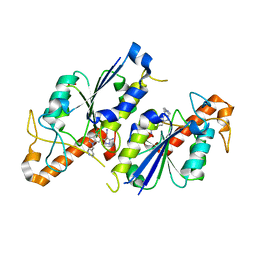 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-(pyridin-4-ylmethyl)indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of Inhibitors against Mycobacterium abscessus tRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
1BET
 
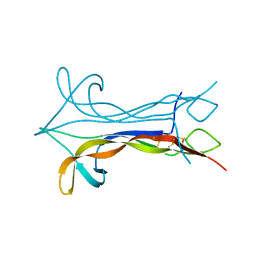 | | NEW PROTEIN FOLD REVEALED BY A 2.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF NERVE GROWTH FACTOR | | Descriptor: | BETA-NERVE GROWTH FACTOR | | Authors: | Mcdonald, N.Q, Lapatto, R, Murray-Rust, J, Gunning, J, Wlodawer, A, Blundell, T.L. | | Deposit date: | 1993-04-08 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor.
Nature, 354, 1991
|
|
1AW8
 
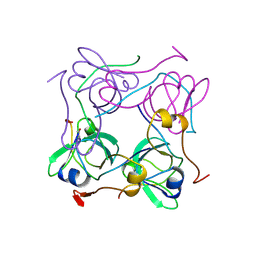 | | PYRUVOYL DEPENDENT ASPARTATE DECARBOXYLASE | | Descriptor: | L-ASPARTATE-ALPHA-DECARBOXYLASE | | Authors: | Albert, A, Dhanaraj, V, Genschel, U, Khan, G, Ramjee, M.K, Pulido, R, Sybanda, B.L, von Delf, F, Witty, M, Blundell, T.L, Smith, A.G, Abell, C. | | Deposit date: | 1997-10-12 | | Release date: | 1998-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aspartate decarboxylase at 2.2 A resolution provides evidence for an ester in protein self-processing.
Nat.Struct.Biol., 5, 1998
|
|
7ZVT
 
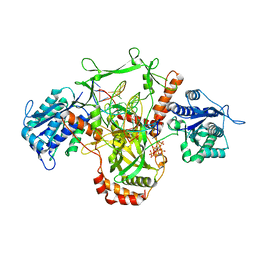 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
1KS9
 
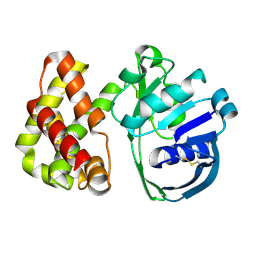 | | Ketopantoate Reductase from Escherichia coli | | Descriptor: | 2-DEHYDROPANTOATE 2-REDUCTASE | | Authors: | Matak-Vinkovic, D, Vinkovic, M, Saldanha, S.A, Ashurst, J.A, von Delft, F, Inoue, T, Miguel, R.N, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism.
Biochemistry, 40, 2001
|
|
7ZWA
 
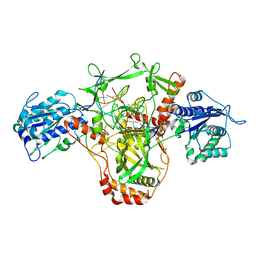 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
1CAQ
 
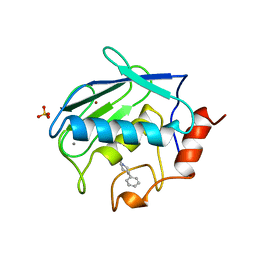 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY | | Descriptor: | 3-(1H-INDOL-3-YL)-2-[4-(4-PHENYL-PIPERIDIN-1-YL)-BENZENESULFONYLAMINO]-PROPIONIC ACID, CALCIUM ION, PROTEIN (STROMELYSIN-1), ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-23 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
1BBS
 
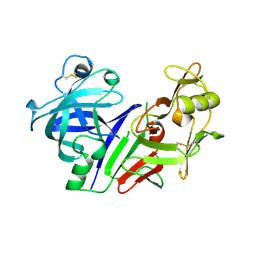 | |
1CIZ
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY | | Descriptor: | 3-(1H-INDOL-3-YL)-2-[4-(4-PHENYL-PIPERIDIN-1-YL)-BENZENESULFONYLAMINO]-PROPIONIC ACID, CALCIUM ION, PROTEIN (STROMELYSIN-1), ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-04-06 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
5MWO
 
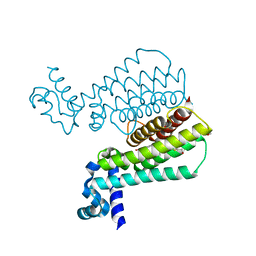 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MXV
 
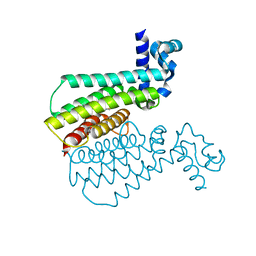 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK1107112A at 1.63A resolution | | Descriptor: | 3-chloranyl-~{N}-(4,5-dihydro-1,3-thiazol-2-yl)-6-fluoranyl-1-benzothiophene-2-carboxamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
7ZYG
 
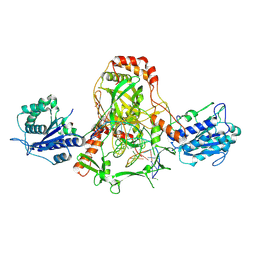 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
5MYL
 
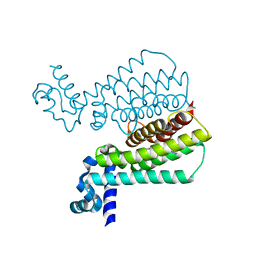 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK1570606A at 1.72A resolution | | Descriptor: | 2-(4-fluorophenyl)-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)ethanamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5MXK
 
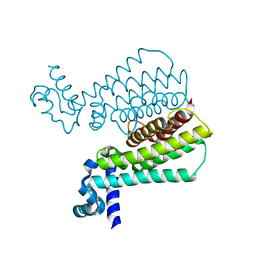 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7G9. | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MYR
 
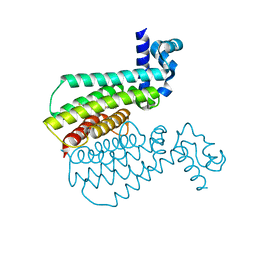 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK735816A at 1.83A resolution | | Descriptor: | 6-fluoranyl-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)-1,3-benzothiazol-2-amine, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5MYW
 
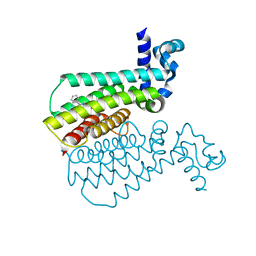 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound SB-435634 at 1.77A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, ~{N}2-(1,3-benzodioxol-5-yl)benzene-1,2-diamine | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5MYM
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK2032710A at 2.28A resolution | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR, [4-(phenylmethyl)piperidin-1-yl]-[1-(5-pyrrol-1-yl-1,3,4-thiadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Mendes, V, Blaszczyk, M, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5MYN
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK445886A at 1.56A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, ~{N}-(5-chloranylpyridin-2-yl)-4-pyridin-2-yl-1,3-thiazol-2-amine | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5MYT
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK921295A at 1.61A resolution | | Descriptor: | 4-methylsulfanyl-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
5MYS
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound GSK920684A at 1.59A resolution | | Descriptor: | 2-(3-fluoranylphenoxy)-~{N}-(4-pyridin-2-yl-1,3-thiazol-2-yl)ethanamide, HTH-type transcriptional regulator EthR | | Authors: | Blaszczyk, M, Mendes, V, Mugumbate, G, Blundell, T.L. | | Deposit date: | 2017-01-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Front Pharmacol, 8, 2017
|
|
1EPO
 
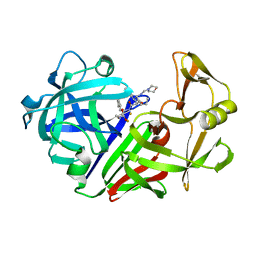 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH CP-81,282 (MOR PHE NLE CHF NME) | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide | | Authors: | Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases.
Protein Sci., 1, 1992
|
|
3COW
 
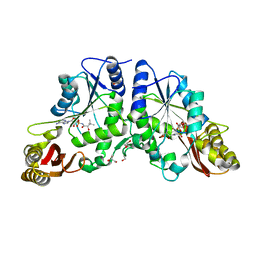 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.8 Ang resolution- in complex with sulphonamide inhibitor 2 | | Descriptor: | 5'-O-{[(2R)-2-hydroxy-3,3-dimethylbutanoyl]sulfamoyl}adenosine, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2008-03-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis pantothenate synthetase by analogues of the reaction intermediate.
Chembiochem, 9, 2008
|
|
3IUB
 
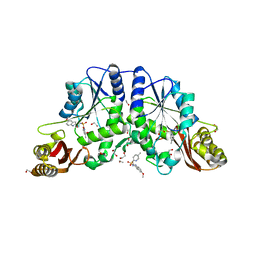 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-Methoxy-N-(5-methylpyridin-2-ylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-methoxy-N-[(5-methylpyridin-2-yl)sulfonyl]-1H-indole-2-carboxamide, ETHANOL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IUE
 
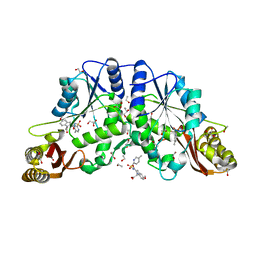 | | Crystal structure of pantothenate synthetase in complex with 2-(5-methoxy-2-(5-Methylpyridin-2-ylsulfonylcarbamoyl)-1H-indol-1-yl) acetic acid | | Descriptor: | (5-methoxy-2-{[(5-methylpyridin-2-yl)sulfonyl]carbamoyl}-1H-indol-1-yl)acetic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L, Wen, S, Hung, A.W, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
