7OTM
 
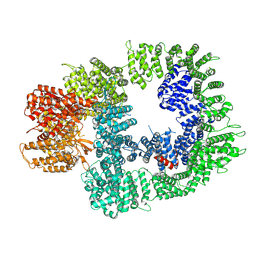 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTY
 
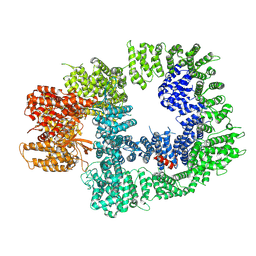 | | DNA-PKcs in complex with M3814 | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTP
 
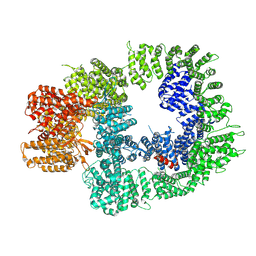 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
5ER1
 
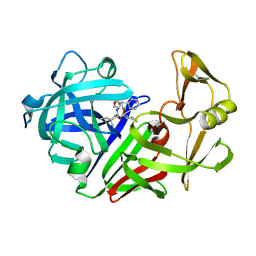 | |
8C1E
 
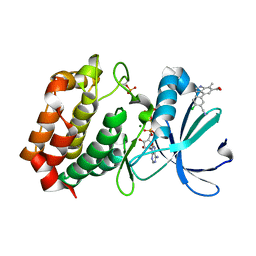 | | Aurora A kinase in complex with TPX2-inhibitor 9 | | Descriptor: | 4-(4-chloranyl-3-cyano-phenyl)-7-methyl-1~{H}-indole-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Selective inhibitors of the Aurora A-TPX2 protein-protein interaction exhibit in vivo efficacy as targeted anti-mitotic agent
To Be Published
|
|
1SMR
 
 | |
4GCR
 
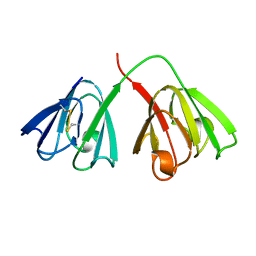 | | STRUCTURE OF THE BOVINE EYE LENS PROTEIN GAMMA-B (GAMMA-II)-CRYSTALLIN AT 1.47 ANGSTROMS | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Slingsby, C, Najmudin, S, Nalini, V, Driessen, H.P.C, Blundell, T.L, Moss, D.S, Lindley, P. | | Deposit date: | 1992-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the bovine eye lens protein gammaB(gammaII)-crystallin at 1.47 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1SGF
 
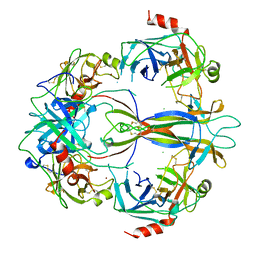 | | CRYSTAL STRUCTURE OF 7S NGF: A COMPLEX OF NERVE GROWTH FACTOR WITH FOUR BINDING PROTEINS (SERINE PROTEINASES) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NERVE GROWTH FACTOR, ... | | Authors: | Bax, B.D.V, Blundell, T.L, Murray-Rust, J, Mcdonald, N.Q. | | Deposit date: | 1997-08-08 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of mouse 7S NGF: a complex of nerve growth factor with four binding proteins.
Structure, 5, 1997
|
|
7ZVT
 
 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZWA
 
 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
2BF9
 
 | | Anisotropic refinement of avian (turkey) pancreatic polypeptide at 0. 99 Angstroms resolution. | | Descriptor: | PANCREATIC HORMONE, ZINC ION | | Authors: | Tickle, I, Glover, I, Pitts, J, Wood, S, Blundell, T.L. | | Deposit date: | 2004-12-06 | | Release date: | 2004-12-08 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Conformational Flexibility in a Small Globular Hormone. X-Ray Analysis of Avian Pancreatic Polypeptide at 0.98 Angstroms Resolution
Biopolymers, 22, 1983
|
|
2WVI
 
 | | Crystal Structure of the N-terminal Domain of BubR1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1 BETA, ... | | Authors: | D'Arcy, S, Davies, O.R, Blundell, T.L, Bolanos-Garcia, V.M. | | Deposit date: | 2009-10-16 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the Molecular Basis of Bubr1 Kinetochore Interactions and Anaphase-Promoting Complex/Cyclosome (Apc/C)-Cdc20 Inhibition
J.Biol.Chem., 285, 2010
|
|
7ZYG
 
 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
4APE
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
1SAC
 
 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
1ER8
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | Endothiapepsin, H-77 | | Authors: | Hemmings, A.M, Veerapandian, B, Szelke, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1989-10-16 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
2QM4
 
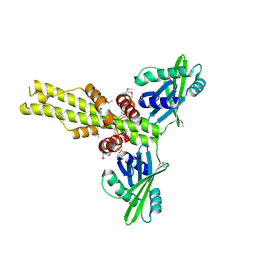 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
2HE0
 
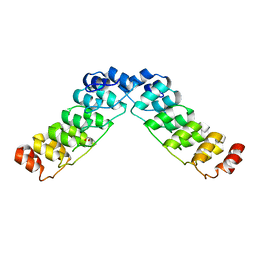 | | Crystal structure of a human Notch1 ankyrin domain mutant | | Descriptor: | 1,2-ETHANEDIOL, Notch1 preproprotein variant | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Martinez Arias, A, Blundell, T.L. | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human Notch1 ankyrin domain mutant
TO BE PUBLISHED
|
|
2JXR
 
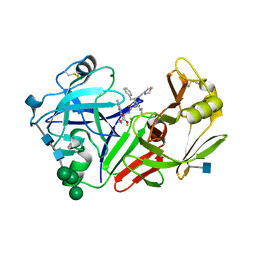 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
1AW8
 
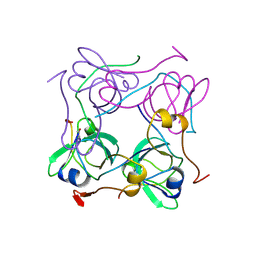 | | PYRUVOYL DEPENDENT ASPARTATE DECARBOXYLASE | | Descriptor: | L-ASPARTATE-ALPHA-DECARBOXYLASE | | Authors: | Albert, A, Dhanaraj, V, Genschel, U, Khan, G, Ramjee, M.K, Pulido, R, Sybanda, B.L, von Delf, F, Witty, M, Blundell, T.L, Smith, A.G, Abell, C. | | Deposit date: | 1997-10-12 | | Release date: | 1998-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aspartate decarboxylase at 2.2 A resolution provides evidence for an ester in protein self-processing.
Nat.Struct.Biol., 5, 1998
|
|
1BBS
 
 | |
1B8Y
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXED WITH NON-PEPTIDE INHIBITORS: IMPLICATIONS FOR INHIBITOR SELECTIVITY | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), SULFATE ION, ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-03 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
1BET
 
 | | NEW PROTEIN FOLD REVEALED BY A 2.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF NERVE GROWTH FACTOR | | Descriptor: | BETA-NERVE GROWTH FACTOR | | Authors: | Mcdonald, N.Q, Lapatto, R, Murray-Rust, J, Gunning, J, Wlodawer, A, Blundell, T.L. | | Deposit date: | 1993-04-08 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor.
Nature, 354, 1991
|
|
3ESL
 
 | |
