5FA2
 
 | | Crystal structure of 426c.TM4deltaV1-3 p120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, THIOCYANATE ION, ... | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
6CHB
 
 | |
6URH
 
 | |
5F7E
 
 | | Crystal structure of germ-line precursor of 3BNC60 Fab | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Sievers, S.A, Scharf, L, Jiang, S, Bjorkman, P.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
7RKS
 
 | | Structure of the SARS-CoV receptor binding domain in complex with the human neutralizing antibody Fab fragment, C118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Antibody Fab Heavy Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKU
 
 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKV
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Fab Heavy Chain, C118 Fab Light Chain, ... | | Authors: | Barnes, C.O, Jette, C.A, Bjorkman, P.J. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
3U7Y
 
 | |
3U7W
 
 | | Crystal structure of NIH45-46 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain, ... | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design.
Science, 334, 2011
|
|
7N3G
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C098 | | Descriptor: | C098 Fab Heavy Chain, C098 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3F
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C080 | | Descriptor: | C080 Fab Heavy Chain, C080 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3H
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C099 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
4JKP
 
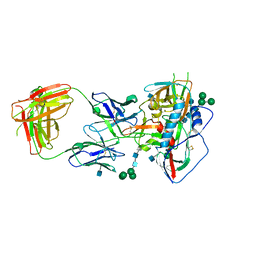 | |
4FQQ
 
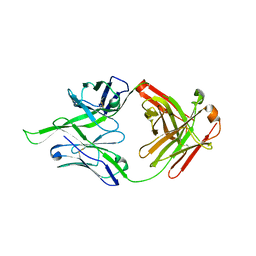 | |
4GW4
 
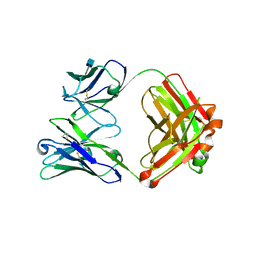 | | Crystal structure of 3BNC60 Fab with P61A mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC60 Fab Heavy-chain, 3BNC60 Fab Light-chain | | Authors: | Diskin, R, Fu, B.Z, Bjorkman, P.J. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Somatic Mutations of the Immunoglobulin Framework Are Generally Required for Broad and Potent HIV-1 Neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
3FRU
 
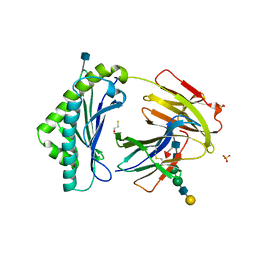 | | NEONATAL FC RECEPTOR, PH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2-MICROGLOBULIN, BETA-MERCAPTOETHANOL, ... | | Authors: | Vaughn, D.E, Burmeister, W.P, Bjorkman, P.J. | | Deposit date: | 1997-12-22 | | Release date: | 1998-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pH-dependent antibody binding by the neonatal Fc receptor.
Structure, 6, 1998
|
|
7R8N
 
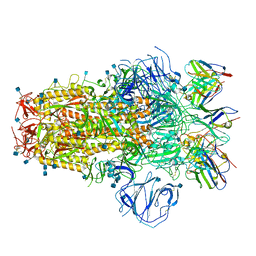 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C051 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8O
 
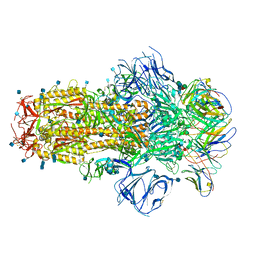 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C548 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3E
 
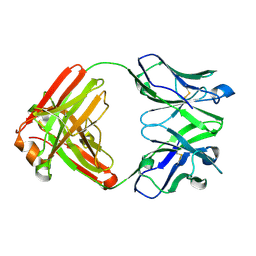 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C032 | | Descriptor: | C032 Fab Heavy Chain, C032 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3I
 
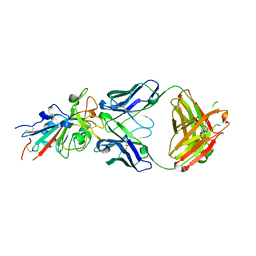 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment C098 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C098 Fab heavy chain, C098 Fab light chain, ... | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
6CH7
 
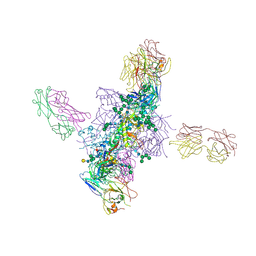 | |
6CH9
 
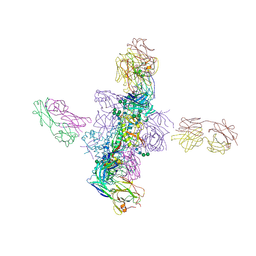 | |
4JDV
 
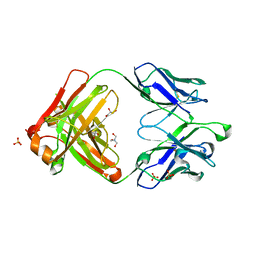 | | Crystal structure of germ-line precursor of NIH45-46 Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Diskin, R, Scharf, L, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5VIC
 
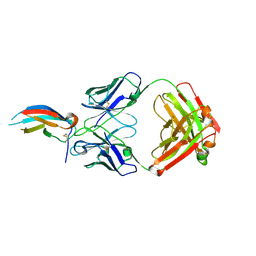 | | Crystal structure of anti-Zika antibody Z004 bound to DENV-1 Envelope protein DIII | | Descriptor: | Dengue 1 Envelope DIII domain, Fab heavy chain, Fab light chain | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
6CH8
 
 | |
