2FEZ
 
 | | Mycobacterium tuberculosis EmbR | | Descriptor: | Probable regulatory protein embR | | Authors: | Futterer, K, Alderwick, L.J, Besra, G.S. | | Deposit date: | 2005-12-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FF4
 
 | |
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
4FEH
 
 | |
4FF6
 
 | | Mycobacterium tuberculosis DprE1 in complex with CT325 - monoclinic crystal form | | Descriptor: | 3-(hydroxyamino)-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FDP
 
 | |
4FDO
 
 | | Mycobacterium tuberculosis DprE1 in complex with CT319 | | Descriptor: | 3-nitro-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, oxidoreductase DprE1 | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-07-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FDN
 
 | |
5DU8
 
 | | Crystal structure of M. tuberculosis EchA6 bound to GSK572A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-[(5-fluoropyridin-2-yl)methyl]-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DUC
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK951A | | Descriptor: | (5R,7S)-N-(1,3-benzodioxol-5-ylmethyl)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DUF
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK729A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxylic acid, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
1SFR
 
 | | Crystal Structure of the Mycobacterium tuberculosis Antigen 85A Protein | | Descriptor: | Antigen 85-A | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-20 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
5DU6
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK059A. | | Descriptor: | (5R,7R)-5-(4-ethylphenyl)-N-(4-fluorobenzyl)-7-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
1R88
 
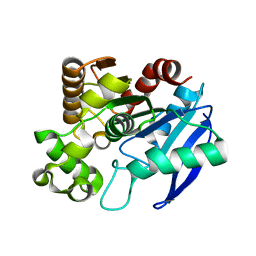 | | The crystal structure of Mycobacterium tuberculosis MPT51 (FbpC1) | | Descriptor: | MPT51/MPB51 antigen | | Authors: | Wilson, R.A, Maughan, W.N, Kremer, L, Besra, G.S, Futterer, K. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structure of Mycobacterium tuberculosis MPT51 (FbpC1) defines a new family of non-catalytic alpha/beta hydrolases.
J.Mol.Biol., 335, 2004
|
|
5DTP
 
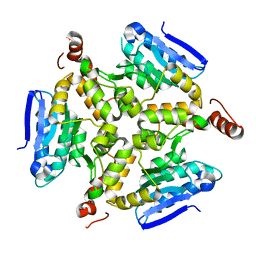 | |
5DU4
 
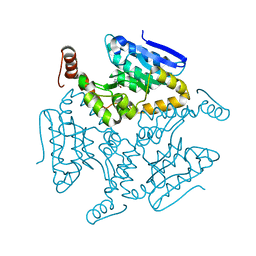 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK366A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-(4-methoxybenzyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DTW
 
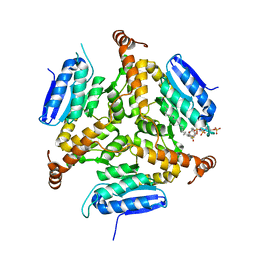 | |
1VA5
 
 | | Antigen 85C with octylthioglucoside in active site | | Descriptor: | Antigen 85-C, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
1T56
 
 | | Crystal structure of TetR family repressor M. tuberculosis EthR | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, EthR repressor, GLYCEROL | | Authors: | Dover, L.G, Corsino, P.E, Daniels, I.R, Cocklin, S.L, Tatituri, V, Besra, G.S, Futterer, K. | | Deposit date: | 2004-05-03 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the TetR/CamR Family Repressor Mycobacterium tuberculosis EthR Implicated in Ethionamide Resistance.
J.Mol.Biol., 340, 2004
|
|
1UQS
 
 | | The Crystal Structure of Human CD1b with a Bound Bacterial Glycolipid | | Descriptor: | BETA-2-MICROGLOBULIN, GLUCOSE MONOMYCOLATE, T-CELL SURFACE GLYCOPROTEIN CD1B | | Authors: | Batuwangala, T, Shepherd, D, Gadola, S.D, Gibson, K.J.C, Zaccai, N.R, Besra, G.S, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2003-10-16 | | Release date: | 2003-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human CD1b with a bound bacterial glycolipid.
J Immunol., 172, 2004
|
|
3OKC
 
 | | Crystal structure of Corynebacterium glutamicum PimB' bound to GDP (orthorhombic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
