6CEU
 
 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
5UKF
 
 | | Crystal Structure of the Human Vaccinia-related Kinase 1 Bound to an Oxindole Inhibitor | | Descriptor: | 4-{[(Z)-(7-oxo-6,7-dihydro-8H-[1,3]thiazolo[5,4-e]indol-8-ylidene)methyl]amino}benzene-1-sulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, Wells, C, Zuercher, W, Willson, T.M, Bountra, C, Edwards, A.M, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-22 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
2XDV
 
 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | Authors: | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-07 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
6CVZ
 
 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
3Q4U
 
 | | Crystal structure of the ACVR1 kinase domain in complex with LDN-193189 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C.D.O, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C.K, Krojer, T, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-24 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
3CYN
 
 | | The structure of human GPX8 | | Descriptor: | GLYCEROL, Probable glutathione peroxidase 8, SULFATE ION | | Authors: | Kavanagh, K.L, Johansson, C, Yue, W.W, Kochan, G, Pike, A.C.W, Murray, J, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GPX8
To be Published
|
|
5LF8
 
 | | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17) | | Descriptor: | ETHYL MERCURY ION, Nucleoside diphosphate-linked moiety X motif 17, PHOSPHATE ION | | Authors: | Mathea, S, Tallant, C, Salah, E, Wang, D, Velupillai, S, Nowak, R, Oerum, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17)
To Be Published
|
|
5HTB
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3353 | | Descriptor: | (3R)-4-amino-3-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}-4-oxobutanoic acid (non-preferred name), (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-11 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3MTF
 
 | | Crystal structure of the ACVR1 kinase in complex with a 2-aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Canning, P, Krojer, T, Vollmar, M, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
3MTN
 
 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
5LXC
 
 | | Crystal structure of DYRK2 in complex with EHT 5372 (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2,4-dichlorophenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
6IBD
 
 | | The Phosphatase and C2 domains of Human SHIP1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Williams, E.P, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5TTF
 
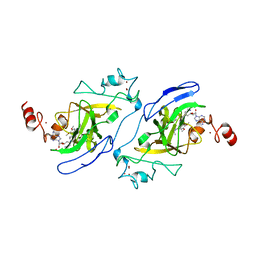 | | Crystal structure of catalytic domain of G9a with MS012 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EHMT2, N4-(1-methylpiperidin-4-yl)-N2-hexyl-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
6I5I
 
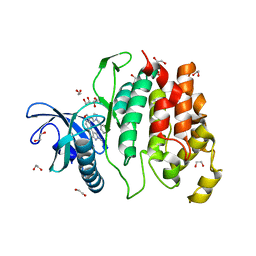 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4XY9
 
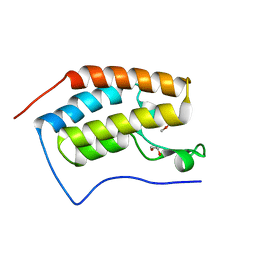 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
5TPX
 
 | | Bromodomain from Plasmodium Faciparum Gcn5, complexed with compound | | Descriptor: | (1S,2S)-N~1~,N~1~-dimethyl-N~2~-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-1-phenylpropane-1,2-diamine, CHLORIDE ION, Histone acetyltransferase GCN5, ... | | Authors: | Lin, Y.H, Hou, C.F.D, MOUSTAKIM, M, DIXON, D.J, Loppnau, P, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, BRENNAN, P.E, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a PCAF Bromodomain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MUF
 
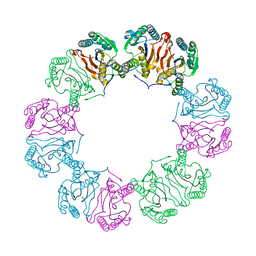 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in its enzymatically active dodecameric form induced by the presence of the N-terminal WDPNWD motif | | Descriptor: | PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
4XYA
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-1-benzofuran-7-yl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
6Y3U
 
 | | Crystal structure of PPARgamma in complex with compound (R)-16 | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
5U4X
 
 | | Coactivator-associated arginine methyltransferase 1 with TP-064 | | Descriptor: | Histone-arginine methyltransferase CARM1, N-methyl-N-[(2-{1-[2-(methylamino)ethyl]piperidin-4-yl}pyridin-4-yl)methyl]-3-phenoxybenzamide, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | DONG, A, ZENG, H, Saikatendu, K.S, STONE, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-06 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma.
Oncotarget, 9, 2018
|
|
6ASD
 
 | | Zinc finger region of human TET1 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3'), Methylcytosine dioxygenase TET1, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
2WWU
 
 | | Crystal structure of the catalytic domain of PHD finger protein 8 | | Descriptor: | ACETATE ION, NICKEL (II) ION, PHD FINGER PROTEIN 8, ... | | Authors: | Yue, W.W, Hozjan, V, Cooper, C, Tumber, A, Krojer, T, Muniz, J, Allerston, C, Salah, E, McDonough, M.A, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Schofield, C.J, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the PHF8 Jumonji domain, an Nepsilon-methyl lysine demethylase.
FEBS Lett., 584, 2010
|
|
5KH3
 
 | | Crystal structure of fragment (3-(5-Chloro-1,3-benzothiazol-2-yl)propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(5-chloranyl-1,3-benzothiazol-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Dong, A, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
2W2I
 
 | | Crystal structure of the human 2-oxoglutarate oxygenase LOC390245 | | Descriptor: | 2-OXOGLUTARATE OXYGENASE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Yue, W.W, Ng, S, Shafqat, N, Ugochukwu, E, McDonough, M, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Schofield, C, Oppermann, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
