6M1W
 
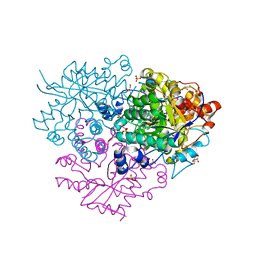 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | Amidohydrolase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4MZ5
 
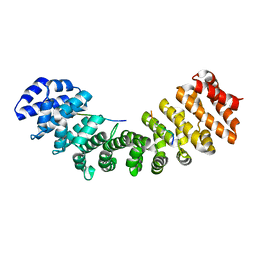 | | Structure of importin-alpha: dUTPase NLS complex | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, Importin subunit alpha-1 | | Authors: | Marfori, M, Rona, G, Vertessy, B.G, Kobe, B. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: structural and mechanistic insights.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6EI4
 
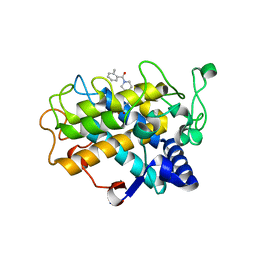 | | Crystal Structure of tyrosinase from Bacillus megaterium with B5N inhibitor in the active site | | Descriptor: | COPPER (II) ION, Tyrosinase, [4-[(4-fluorophenyl)methyl]piperazin-1-yl]-(2-methylphenyl)methanone | | Authors: | Deri, B, Gitto, R, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-09-17 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Tyrosinase: Development and Structural Insights of Novel Inhibitors Bearing Arylpiperidine and Arylpiperazine Fragments.
J. Med. Chem., 61, 2018
|
|
4N4J
 
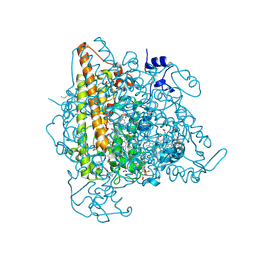 | | Kuenenia stuttgartiensis hydroxylamine oxidoreductase | | Descriptor: | 1-[(4-cyclohexylbutanoyl)(2-hydroxyethyl)amino]-1-deoxy-D-glucitol, HEME C, PHOSPHATE ION, ... | | Authors: | Maalcke, W.J, Dietl, A, Marritt, S.J, Butt, J.N, Jetten, M.S.M, Keltjens, J.T, Barends, T.R.M.B, Kartal, B. | | Deposit date: | 2013-10-08 | | Release date: | 2013-12-11 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
J.Biol.Chem., 289, 2014
|
|
4N3S
 
 | |
6EKP
 
 | | Tryptophan Repressor TrpR from E.coli variant T44L T81M S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, 3,6,9,12,15-PENTAOXAHEPTADECANE, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-26 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
4N68
 
 | | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804] | | Descriptor: | Contactin-5, SULFATE ION | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804]
to be published
|
|
6ME2
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
4N8P
 
 | | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Uncharacterized protein | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711]
to be published
|
|
6ELF
 
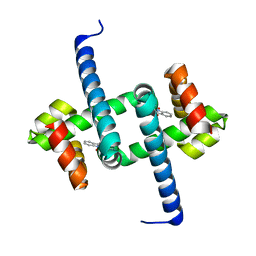 | | Tryptophan Repressor TrpR from E.coli variant M42F T44L T81I S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
6LSF
 
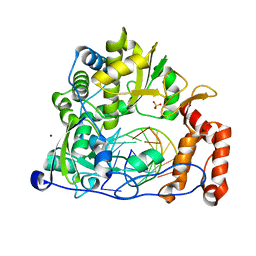 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6RA/C2S6RB form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
6EMW
 
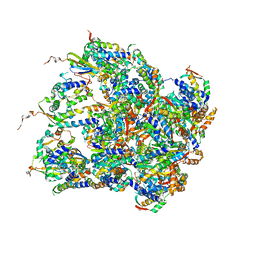 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6MDS
 
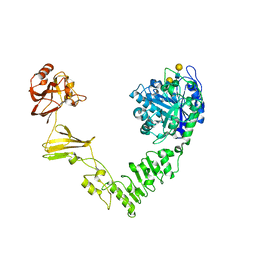 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with complex biantennary glycan | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Klontz, E.H, Trastoy, B, Orwenyo, J, Wang, L.X, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
6ME4
 
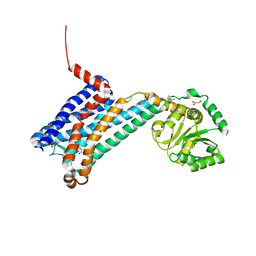 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6LNB
 
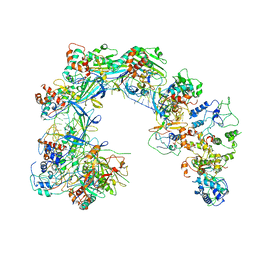 | | CryoEM structure of Cascade-TniQ-dsDNA complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6ELY
 
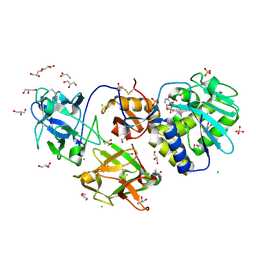 | | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.84 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-N-Furfurylcytosine, ... | | Authors: | Ahmad, M.S, Rasheed, S, Falke, S, Khaliq, B, Perbandt, M, Choudhary, M.I, Markiewicz, W.T, Barciszewski, J, Betzel, C. | | Deposit date: | 2017-09-30 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.85 angstrom Resolution.
Med Chem, 14, 2018
|
|
6EHB
 
 | |
6EM0
 
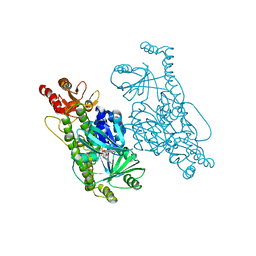 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase M321A from Pseudomonas azelaica | | Descriptor: | 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Deri, B, Bregman-Cohen, A, Pazy Benhar, Y, Fishman, A. | | Deposit date: | 2017-10-01 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Altering 2-Hydroxybiphenyl 3-Monooxygenase Regioselectivity by Protein Engineering for the Production of a New Antioxidant.
Chembiochem, 19, 2018
|
|
6LMK
 
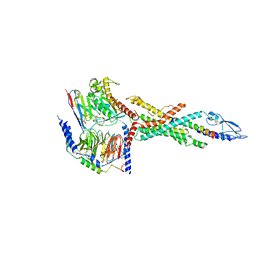 | | Cryo-EM structure of the human glucagon receptor in complex with Gs | | Descriptor: | Glucagon, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qiao, A, Han, S, Tai, L, Sun, F, Zhao, Q, Wu, B. | | Deposit date: | 2019-12-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of Gsand Girecognition by the human glucagon receptor.
Science, 367, 2020
|
|
6M2I
 
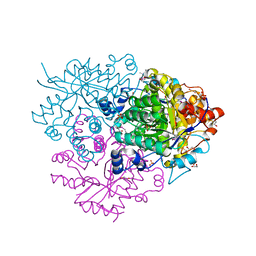 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
6LRZ
 
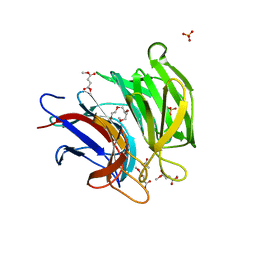 | | Crystal structure of Keap1 in complex with dimethyl fumarate (DMF) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, Keap1-DC, ... | | Authors: | Padmanabhan, B, Unni, S, Deshmukh, P. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Febs J., 288, 2021
|
|
6LTJ
 
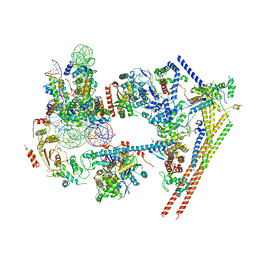 | | Structure of nucleosome-bound human BAF complex | | Descriptor: | AT-rich interactive domain-containing protein 1A, Actin, cytoplasmic 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
6LZE
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11a | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhang, Y, Jing, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6MEO
 
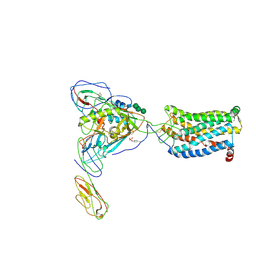 | | Structural basis of coreceptor recognition by HIV-1 envelope spike | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaik, M.M, Chen, B. | | Deposit date: | 2018-09-06 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of coreceptor recognition by HIV-1 envelope spike.
Nature, 565, 2018
|
|
