8UUY
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with inhibitor Jun12129 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UVM
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with covalent inhibitor Jun11313 | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Ansari, A, Tan, B, Chopra, A, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-03 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUV
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12197 | | Descriptor: | CHLORIDE ION, N-{(1R)-1-[(3P,5P)-3-(1-cyclopropyl-1H-pyrazol-4-yl)-5-(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, Papain-like protease nsp3, ... | | Authors: | Ansari, A, Tan, B, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUH
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12199 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-[(1R)-1-{(3M,5P)-3-(1-methyl-1H-pyrazol-4-yl)-5-[1-(propan-2-yl)-1H-pyrazol-4-yl]phenyl}ethyl]benzamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUU
 
 | | SARS-Cov-2 papain-like protease (PLpro) with inhibitor Jun12162 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUW
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12145 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-2-methyl-N-{(1R)-1-[(3P,5M)-3-(1-methyl-1H-pyrazol-4-yl)-5-(1,3-thiazol-5-yl)phenyl]ethyl}benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
1ZYR
 
 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
6HAK
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with a double stranded RNA represents the RT transcription initiation complex prior to nucleotide incorporation | | Descriptor: | Gag-Pol polyprotein, MAGNESIUM ION, RNA (5'-R(P*AP*GP*UP*GP*GP*CP*GP*GP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2018-08-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2CW0
 
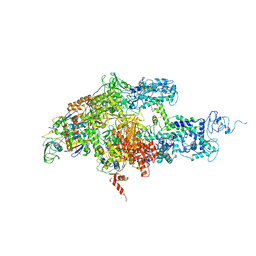 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
4QAG
 
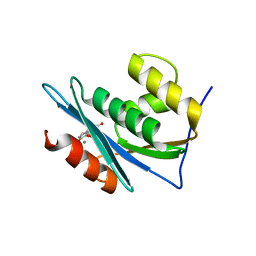 | | Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNASE H domain of HIV-1 reverse transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Himmel, D.M, Ho, W.C, Arnold, E. | | Deposit date: | 2014-05-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs.
J.Mol.Biol., 426, 2014
|
|
1S6P
 
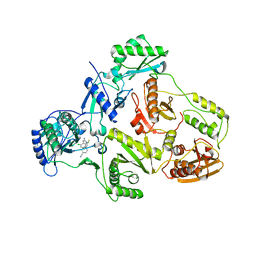 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R100943 | | Descriptor: | 1-(4-CYANO-PHENYL)-3-[2-(2,6-DICHLORO-PHENYL)-1-IMINO-ETHYL]-THIOUREA, MAGNESIUM ION, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
5KRS
 
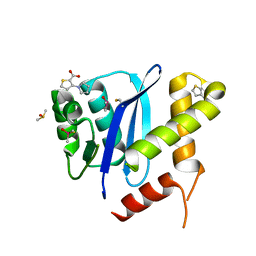 | | HIV-1 Integrase Catalytic Core Domain in Complex with an Allosteric Inhibitor, 3-(1H-pyrrol-1-yl)-2-thiophenecarboxylic acid | | Descriptor: | 3-pyrrol-1-ylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, Integrase | | Authors: | Patel, D, Bauman, J.D, Arnold, E. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-28 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
J.Biol.Chem., 291, 2016
|
|
5KRT
 
 | |
1S6Q
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9E
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1SUQ
 
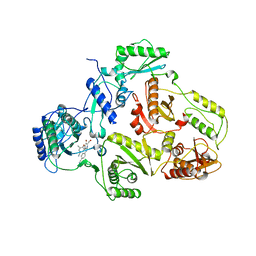 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | Descriptor: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1SV5
 
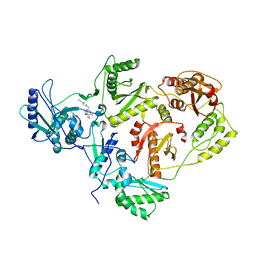 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-27 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1J5O
 
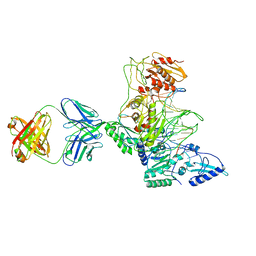 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6UL5
 
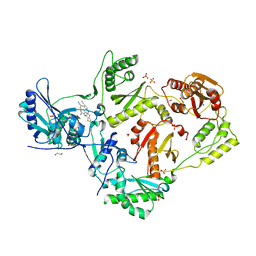 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Pilch, A, Arnold, E. | | Deposit date: | 2019-10-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
8DX2
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-amino-3-bromopyridine at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 3-bromopyridin-4-amine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXK
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-bromopyrazole at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXJ
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 1-N-methyl-4-(trifluoromethyl)benzene-1,2-diamine at the NNRTI adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXI
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment [1-(4-fluorophenyl)-5-methyl-1H-pyrazol-4-yl]methanol at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DXG
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 5-(trifluoromethyl)pyridin-2-ol at W24 site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, 5-(trifluoromethyl)pyridin-2-one, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
