1XHO
 
 | | Chorismate mutase from Clostridium thermocellum Cth-682 | | Descriptor: | Chorismate mutase, UNKNOWN ATOM OR ION | | Authors: | Xu, H, Chen, L, Lee, D, Habel, J.E, Nguyen, J, Chang, S.-H, Kataeva, I, Chang, J, Zhao, M, Yang, H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Away from the edge II: in-house Se-SAS phasing with chromium radiation.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2KBF
 
 | |
3RY7
 
 | | Crystal Structure of Sa239 | | Descriptor: | GLYCEROL, Ribokinase | | Authors: | Li, J, Wu, M, Wang, L, Zang, J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Sa239 reveals the structural basis for the activation of ribokinase by monovalent cations.
J.Struct.Biol., 177, 2012
|
|
3U50
 
 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 OB-C | | Descriptor: | Telomerase-associated protein 82, ZINC ION | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U82
 
 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | Descriptor: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.164 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
2D28
 
 | | Structure of the N-terminal domain of XpsE (crystal form P43212) | | Descriptor: | CACODYLATE ION, type II secretion ATPase XpsE | | Authors: | Chen, Y, Shiue, S.-J, Huang, C.-W, Chang, J.-L, Chien, Y.-L, Hu, N.-T, Chan, N.-L. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the XpsE N-Terminal Domain, an Essential Component of the Xanthomonas campestris Type II Secretion System
J.Biol.Chem., 280, 2005
|
|
4HK4
 
 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
6JRL
 
 | | Crystal structure of Drosophila alpha methyldopa-resistant protein/3,4-dihydroxyphenylacetaldehyde synthase | | Descriptor: | 3,4-dihydroxyphenylacetaldehyde synthase | | Authors: | Wei, S, Vavrick, C.J, Guan, H, Liao, C, Robinson, H, Liang, J, Wang, D, Han, Q, Li, J. | | Deposit date: | 2019-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the bifunctional mechanism of Drosophila alpha methyldopa-resistant protein/3,4-dihydroxyphenylacetaldehyde synthase
To Be Published
|
|
7L7W
 
 | | Crystal structure of Arabidopsis NRG1.1 CC-R domain K94E/K96E mutant | | Descriptor: | NICKEL (II) ION, Probable disease resistance protein At5g66900 | | Authors: | Walton, W.G, Wan, L, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Plant "helper" immune receptors are Ca 2+ -permeable nonselective cation channels.
Science, 373, 2021
|
|
7L99
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1302 | | Descriptor: | Bromodomain testis-specific protein, N-[3-(acetylamino)-4-methylphenyl]-3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carboxamide, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Sharma, R, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4QB9
 
 | | Crystal structure of Mycobacterium smegmatis Eis in complex with paromomycin | | Descriptor: | Enhanced intracellular survival protein, PAROMOMYCIN, SULFATE ION | | Authors: | Kim, K.H, Ahn, D.R, Yoon, H.J, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structure of Mycobacterium smegmatis Eis in complex with paromomycin.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7L9A
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8PYI
 
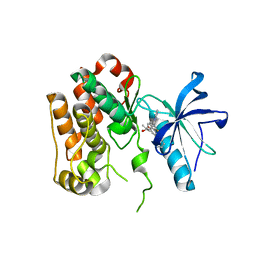 | | Human IGF1R with inhibitor 6 | | Descriptor: | 3-[8-azanyl-1-(4-ethoxy-8-fluoranyl-2-phenyl-quinolin-7-yl)imidazo[1,5-a]pyrazin-3-yl]-1-methyl-cyclobutan-1-ol, Insulin-like growth factor 1 receptor beta chain | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Human IGF1R with inhibitor 6
To Be Published
|
|
8PYJ
 
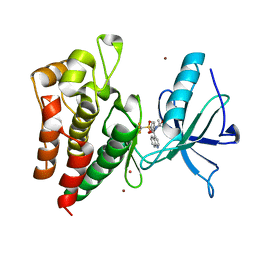 | | Human IGF1R with inhibitor 8 | | Descriptor: | 5,5-dimethyl-1-(quinolin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Human IGF1R with inhibitor 8
To Be Published
|
|
8PYK
 
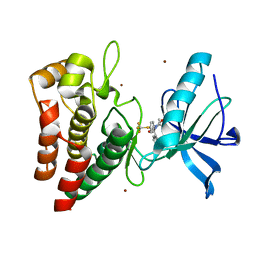 | | Human IGF1R with inhibitor 47 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, Insulin-like growth factor 1 receptor beta chain, NICKEL (II) ION | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human IGF1R with inhibitor 47
To Be Published
|
|
8PYN
 
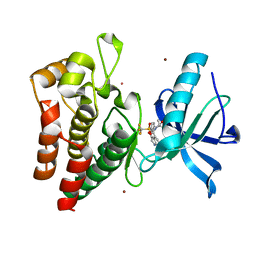 | | Human IGF1R with inhibitor 56 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Human IGF1R with inhibitor 56
To Be Published
|
|
8PYL
 
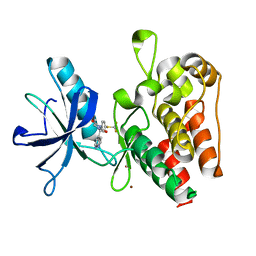 | | Human IGF1R with inhibitor 53 | | Descriptor: | (5S)-5-methyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Human IGF1R with inhibitor 53
To Be Published
|
|
5WS3
 
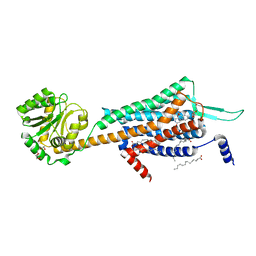 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
8PYM
 
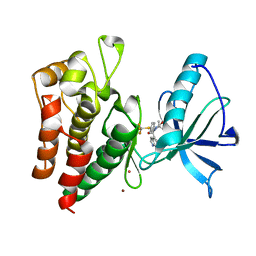 | | Human IGF1R with inhibitor 54 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Human IGF1R with inhibitor 54
To Be Published
|
|
4HPW
 
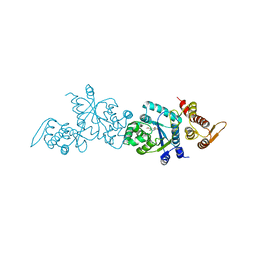 | | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine | | Descriptor: | 3-methoxy-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine
To be Published
|
|
7C39
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with methylated L-fucose | | Descriptor: | AoflcA, CITRIC ACID, GLYCEROL, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7CLV
 
 | | Solution structure of mitochondrial Tim23 channel in complex with a signaling peptide | | Descriptor: | COX4 isoform 1, TIM23 isoform 1 | | Authors: | Zhou, S, Ruan, M.S, Li, Y.Y, Yang, J, Richter, C, Schwalbe, H, Shen, B, Wang, J.F. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide.
Cell Res., 31, 2021
|
|
7C3D
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with D-arabinose | | Descriptor: | AofleA, GLYCEROL, alpha-D-arabinopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
7C38
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with L-fucose | | Descriptor: | AofleA, GLYCEROL, alpha-L-fucopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
