3DH3
 
 | | Crystal Structure of RluF in complex with a 22 nucleotide RNA substrate | | Descriptor: | Ribosomal large subunit pseudouridine synthase F, stem loop fragment of E. Coli 23S RNA | | Authors: | Alian, A, DeGiovanni, A, Stroud, R.M, Finer-Moore, J.S. | | Deposit date: | 2008-06-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an RluF-RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome.
J.Mol.Biol., 388, 2009
|
|
2G9X
 
 | |
5AB0
 
 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
3DDQ
 
 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor roscovitine | | Descriptor: | Cell division protein kinase 2, Cyclin-A2, MONOTHIOGLYCEROL, ... | | Authors: | Echalier, A, Endicott, J.A. | | Deposit date: | 2008-06-06 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CR8, a potent and selective, roscovitine-derived inhibitor of cyclin-dependent kinases.
Oncogene, 27, 2008
|
|
5AB2
 
 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
7ZYF
 
 | | Insulin regulated aminopeptidase (IRAP) in complex with a nanomolar alpha hydroxy beta amino acid based inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P, Papakyriakou, A. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
5C97
 
 | | Insulin regulated aminopeptidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucyl-cystinyl aminopeptidase, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
5MJ6
 
 | | Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Mpakali, A, Stratikos, E, Saridakis, E, Giastas, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Ligand-Induced Conformational Change of Insulin-Regulated Aminopeptidase: Insights on Catalytic Mechanism and Active Site Plasticity.
J. Med. Chem., 60, 2017
|
|
5LMK
 
 | | Structure of phopsho-CDK2-cyclin A in complex with an ATP-competitive inhibitor | | Descriptor: | 4-[4-[3-bromanyl-7-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-5-yl]phenyl]benzamide, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Explicit treatment of active-site waters enhances quantum mechanical/implicit solvent scoring: Inhibition of CDK2 by new pyrazolo[1,5-a]pyrimidines.
Eur J Med Chem, 126, 2016
|
|
3BHT
 
 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor meriolin 3 | | Descriptor: | 4-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
3BHV
 
 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor variolin B | | Descriptor: | 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
3BHU
 
 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor meriolin 5 | | Descriptor: | 4-(4-propoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
8A8S
 
 | |
8AA8
 
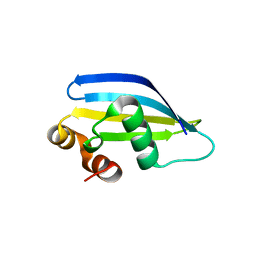 | |
5ZEF
 
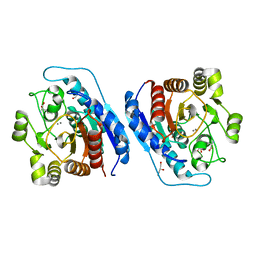 | | Crystal structure of Entamoeba histolytica Arginase in complex with L- Norvaline at 2.01 A | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL, ... | | Authors: | Malik, A, Dalal, V, Ankri, S, Tomar, S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules.
Febs J., 286, 2019
|
|
5ZEH
 
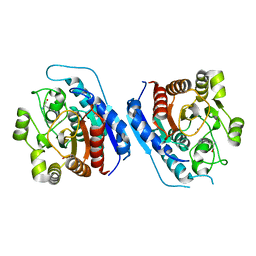 | | Crystal structure of Entamoeba histolytica Arginase in complex with L- Ornithine at 2.35 A | | Descriptor: | 1,2-ETHANEDIOL, Arginase, L-ornithine, ... | | Authors: | Malik, A, Dalal, V, Ankri, S, Tomar, S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules.
Febs J., 286, 2019
|
|
5ZEE
 
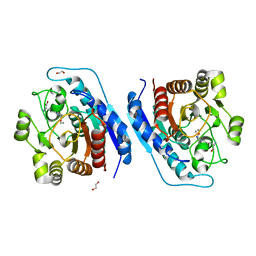 | | Crystal structure of Entamoeba histolytica Arginase in complex with N(omega)-hydroxy-L-arginine (NOHA) at 1.74 A | | Descriptor: | 1,2-ETHANEDIOL, Arginase, MANGANESE (II) ION, ... | | Authors: | Malik, A, Dalal, V, Ankri, S, Tomar, S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules.
Febs J., 286, 2019
|
|
4Z7I
 
 | | Crystal structure of insulin regulated aminopeptidase in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DG025 transition-state analogue enzyme inhibitor, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-04-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
3BT7
 
 | |
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
4EOR
 
 | | Thr 160 phosphorylated CDK2 WT - human cyclin A3 complex with the inhibitor NU6102 | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EON
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, Q131E - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOL
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOQ
 
 | | Thr 160 phosphorylated CDK2 WT - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
1FRQ
 
 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
