9C3T
 
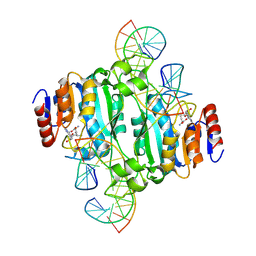 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrate containing GTAAAC as recognition sequence | | Descriptor: | DNA1, DNA2, Methyltransferase, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2024-06-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
9C3U
 
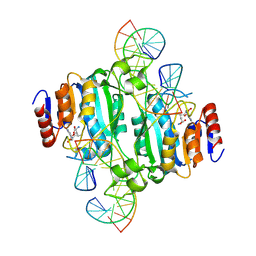 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrate containing GTTTAC as recognition sequence | | Descriptor: | DNA1, DNA2, Methyltransferase, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2024-06-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
7TW9
 
 | |
8TLT
 
 | | Cryo-EM structure of Rev1(deltaN)-Polzeta-DNA-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*TP*AP*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Malik, R, Aggarwal, A.K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4PTF
 
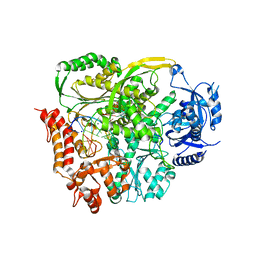 | | Ternary crystal structure of yeast DNA polymerase epsilon with template G | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', ... | | Authors: | Jain, R, Rajashankar, K.R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Crystal Structure of Yeast DNA Polymerase epsilon Catalytic Domain.
Plos One, 9, 2014
|
|
8TLQ
 
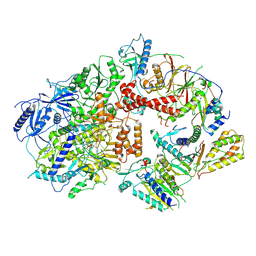 | | Cryo-EM structure of the Rev1-Polzeta-DNA-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Malik, R, Aggarwal, A.K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7TW8
 
 | |
7TW7
 
 | |
1IF1
 
 | | INTERFERON REGULATORY FACTOR 1 (IRF-1) COMPLEX WITH DNA | | Descriptor: | DNA (26-MER), PROTEIN (INTERFERON REGULATORY FACTOR 1) | | Authors: | Escalante, C.R, Yie, J, Thanos, D, Aggarwal, A. | | Deposit date: | 1997-09-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of IRF-1 with bound DNA reveals determinants of interferon regulation.
Nature, 391, 1998
|
|
6P1H
 
 | | Cryo-EM Structure of DNA Polymerase Delta Holoenzyme | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Jain, R, Rice, W, Aggarwal, A.K. | | Deposit date: | 2019-05-19 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure and dynamics of eukaryotic DNA polymerase delta holoenzyme.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8FMF
 
 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (1 tetramer in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FM1
 
 | |
8FMG
 
 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (3 tetramers in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, MAGNESIUM ION, SAVED domain-containing protein, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FMH
 
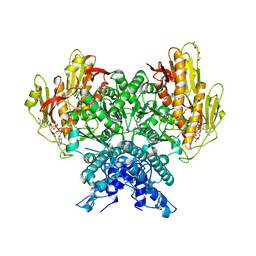 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-dGAMP ligand (2 tetramers in the AU) | | Descriptor: | 3'2'-cGAMP, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6PZ3
 
 | |
8FRK
 
 | |
8FRJ
 
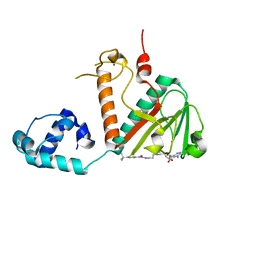 | | Structure of nsp14 N7-MethylTransferase domain fused with TELSAM bound to SGC0946 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Transcription factor ETV6,Guanine-N7 methyltransferase nsp14 chimera, ZINC ION | | Authors: | Kottur, J, Aggarwal, A.K. | | Deposit date: | 2023-01-07 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of SARS-CoV-2 N7-methyltransferase with DOT1L and PRMT7 inhibitors provide a platform for new antivirals.
Plos Pathog., 19, 2023
|
|
3BSB
 
 | | Crystal Structure of Human Pumilio1 in Complex with CyclinB reverse RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*AP*UP*GP*UP*U)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-23 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
8URK
 
 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrates | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA1, DNA2, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2023-10-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
6Q02
 
 | |
8U7B
 
 | |
8U72
 
 | | Cryo-EM structure of the SPARTA oligomer with guide RNA and target DNA | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*C)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Malik, R, Kottur, J, Aggarwal, A.K. | | Deposit date: | 2023-09-14 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Nucleic acid mediated activation of a short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
3BSX
 
 | | Crystal Structure of Human Pumilio 1 in complex with Puf5 RNA | | Descriptor: | 5'-R(*UP*UP*GP*UP*AP*AP*UP*AP*UP*UP*A)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
8VCC
 
 | |
5HR4
 
 | | Structure of Type IIL restriction-modification enzyme MmeI in complex with DNA has implications for engineering of new specificities | | Descriptor: | CALCIUM ION, DNA (5'-D(P*GP*TP*TP*AP*TP*GP*TP*CP*GP*GP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*CP*CP*GP*AP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Callahan, S.J, Luyten, Y.A, Gupta, Y.K, Wilson, G.G, Roberts, R.J, Morgan, R.D, Aggarwal, A.K. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5964 Å) | | Cite: | Structure of Type IIL Restriction-Modification Enzyme MmeI in Complex with DNA Has Implications for Engineering New Specificities.
Plos Biol., 14, 2016
|
|
