6FO0
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial compound GSK932121 | | Descriptor: | 3-chloro-6-(hydroxymethyl)-2-methyl-5-{4-[3-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, Chain I/V, Cytochrome b, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6NI3
 
 | | B2V2R-Gs protein subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Endolysin,Beta-2 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NQU
 
 | | Human LSD1 in complex with GSK2879552 | | Descriptor: | Lysine-specific histone demethylase 1A, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S})-5-[(9~{S},11~{R})-15,16-dimethyl-11-oxidanyl-5,7-bis(oxidanylidene)-9-phenyl-2,4,6,12-tetrazabicyclo[11.4.0]heptadeca-1(17),13,15-trien-2-yl]-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Tan, A.H.Y, Tu, W, McCuaig, R, Donovan, T, Tsimbalyuk, S, Forwood, J.K, Rao, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lysine-Specific Histone Demethylase 1A Regulates Macrophage Polarization and Checkpoint Molecules in the Tumor Microenvironment of Triple-Negative Breast Cancer.
Front Immunol, 10, 2019
|
|
6GQ3
 
 | | Human asparagine synthetase (ASNS) in complex with 6-diazo-5-oxo-L-norleucine (DON) at 1.85 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-OXO-L-NORLEUCINE, ... | | Authors: | Zhu, W, Radadiya, A, Bisson, C, Jin, Y, Nordin, B.E, Imasaki, T, Wenzel, S, Sedelnikova, S.E, Berry, A.H, Nomanbhoy, T.K, Kozarich, J.W, Takagi, Y, Rice, D.W, Richards, N.G.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity.
Commun Biol, 2, 2019
|
|
6NI2
 
 | | Stabilized beta-arrestin 1-V2T subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NQM
 
 | | Crystal structure of Human LSD1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A | | Authors: | Tan, A.H.Y, Tu, W, McCuaig, R, Donovan, T, Tsimbalyuk, S, Forwood, J.K, Rao, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lysine-Specific Histone Demethylase 1A Regulates Macrophage Polarization and Checkpoint Molecules in the Tumor Microenvironment of Triple-Negative Breast Cancer.
Front Immunol, 10, 2019
|
|
6FO2
 
 | | CryoEM structure of bovine cytochrome bc1 with no ligand bound | | Descriptor: | Cytochrome b, Cytochrome b-c1 complex subunit 1, mitochondrial, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6FPN
 
 | | Lytic transglycosylase in action | | Descriptor: | Putative soluble lytic murein transglycosylase, SULFATE ION | | Authors: | Williams, A.H, Hoauz, A, Boneca, I.G. | | Deposit date: | 2018-02-11 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A step-by-stepin crystalloguide to bond cleavage and 1,6-anhydro-sugar product synthesis by a peptidoglycan-degrading lytic transglycosylase.
J. Biol. Chem., 293, 2018
|
|
6FO6
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial inhibitor SCR0911 | | Descriptor: | 7-methoxy-3-methyl-2-[5-[4-(trifluoromethyloxy)phenyl]pyridin-3-yl]quinolin-4-ol, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6EF1
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6H5F
 
 | | LtgA disordered Helix | | Descriptor: | Putative soluble lytic murein transglycosylase | | Authors: | Williams, A.H. | | Deposit date: | 2018-07-24 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Defective lytic transglycosylase disrupts cell morphogenesis by hindering cell wall de-O-acetylation inNeisseria meningitidis.
Elife, 9, 2020
|
|
5AYR
 
 | | The crystal structure of SAUGI/human UDG complex | | Descriptor: | MAGNESIUM ION, Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
5AYS
 
 | | Crystal structure of SAUGI/HSV UDG complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
5XNR
 
 | | Truncated AlyQ with CBM32 and alginate lyase domains | | Descriptor: | ACETATE ION, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2.
Sci Rep, 7, 2017
|
|
2Z9L
 
 | |
2ZED
 
 | | Crystal structure of the human glutaminyl cyclase mutant S160A at 1.7 angstrom resolution | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Wang, Y.R, Chang, E.C, Chou, T.L, Wang, A.H. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved hydrogen-bond network in the catalytic centre of animal glutaminyl cyclases is critical for catalysis.
Biochem.J., 411, 2008
|
|
2Z8V
 
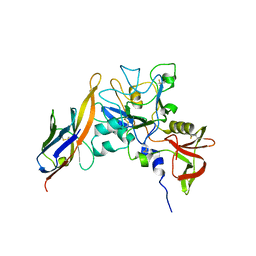 | | Structure of an IgNAR-AMA1 complex | | Descriptor: | Apical membrane antigen 1, New antigen receptor variable domain | | Authors: | Streltsov, V.A, Henderson, K.A, Batchelor, A.H, Coley, A.M, Nuttall, S.D. | | Deposit date: | 2007-09-11 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of an IgNAR-AMA1 Complex: Targeting a Conserved Hydrophobic Cleft Broadens Malarial Strain Recognition
Structure, 15, 2007
|
|
2Z9G
 
 | | Complex structure of SARS-CoV 3C-like protease with PMA | | Descriptor: | 3C-like proteinase, BENZENE, MERCURY (II) ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2ZCS
 
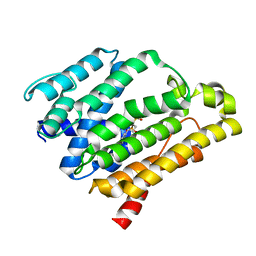 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-700 | | Descriptor: | Dehydrosqualene synthase, tripotassium (1R)-4-biphenyl-4-yl-1-phosphonatobutane-1-sulfonate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
2ZEH
 
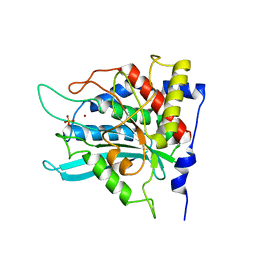 | | Crystal structure of the human glutaminyl cyclase mutant E201Q at 1.8 angstrom resolution | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Wang, Y.R, Chang, E.C, Chou, T.L, Wang, A.H. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved hydrogen-bond network in the catalytic centre of animal glutaminyl cyclases is critical for catalysis.
Biochem.J., 411, 2008
|
|
2Z94
 
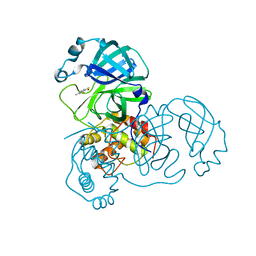 | | Complex structure of SARS-CoV 3C-like protease with TDT | | Descriptor: | 4-methylbenzene-1,2-dithiol, Replicase polyprotein 1ab, ZINC ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-17 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors
Febs Lett., 581, 2007
|
|
5XB7
 
 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
2Z9K
 
 | | Complex structure of SARS-CoV 3C-like protease with JMF1600 | | Descriptor: | (dimethylamino)(hydroxy)zinc', 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
5XHV
 
 | | Crystal Structure Of Fab S40 In Complex With Influenza Hemagglutinin, HA1 subunit. | | Descriptor: | Hemagglutinin, S40 heavy chain, S40 light chain | | Authors: | Lee, C.C, Wang, A.H.J, Yu, C.M, Yang, A.S. | | Deposit date: | 2017-04-24 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries.
Sci Rep, 7, 2017
|
|
2ZEN
 
 | | Crystal structure of the human glutaminyl cyclase mutant D305A at 1.78 angstrom resolution | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Wang, Y.R, Chang, E.C, Chou, T.L, Wang, A.H. | | Deposit date: | 2007-12-13 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A conserved hydrogen-bond network in the catalytic centre of animal glutaminyl cyclases is critical for catalysis.
Biochem.J., 411, 2008
|
|
