2VZE
 
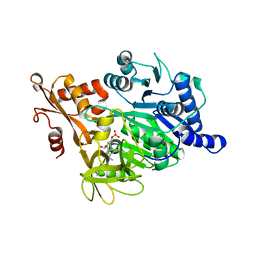 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with AMP | | Descriptor: | ACYL-COENZYME A SYNTHETASE ACSM2A, MITOCHONDRIAL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Yue, W.W, Kochan, G.T, Pilka, E.S, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wikstrom, M, Bountra, C, Oppermann, U. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Snapshots for the Conformation- Dependent Catalysis by Human Medium-Chain Acyl- Coenzyme a Synthetase Acsm2A.
J.Mol.Biol., 388, 2009
|
|
6VV2
 
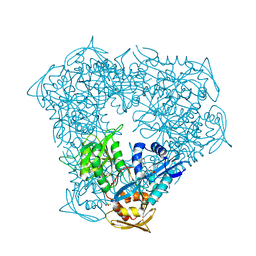 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1348 | | Descriptor: | 2-{[3-(piperidin-1-yl)propyl]sulfanyl}-6,7,8,9-tetrahydro-5H-cyclohepta[4,5]thieno[2,3-d]pyrimidin-4-amine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
1K1G
 
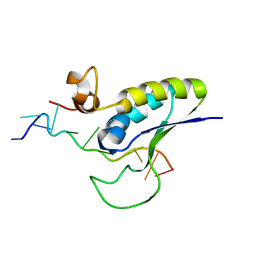 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
5KCU
 
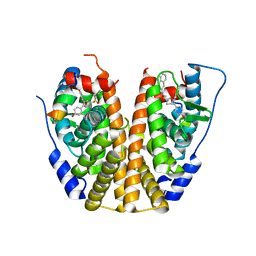 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
6PJC
 
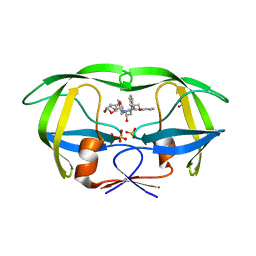 | | HIV-1 Protease NL4-3 WT in Complex with LR4-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-5-{[(2,6-dimethylphenoxy)acetyl]amino}-4-hydroxy-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.965 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
5KGE
 
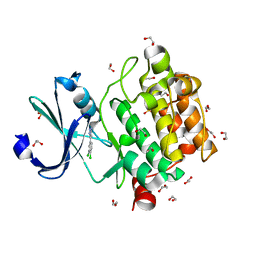 | |
2II3
 
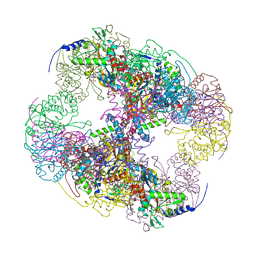 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Oxidized Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2ILK
 
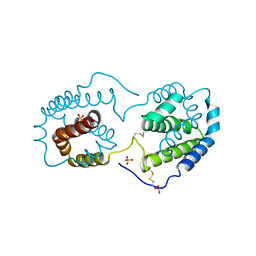 | |
1K2F
 
 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
4V27
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Hakki, Z, Bellmaine, S, Thompson, A.J, Speciale, G, Davies, G.J, Williams, S.J. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Dissection of the Endo-Alpha-1,2-Mannanase Activity of Bacterial Gh99 Glycoside Hydrolases from Bacteroides Spp.
Chemistry, 21, 2015
|
|
3EAH
 
 | | Structure of inhibited human eNOS oxygenase domain | | Descriptor: | (3S,5E)-3-propyl-3,4-dihydrothieno[2,3-f][1,4]oxazepin-5(2H)-imine, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
4IKR
 
 | | Crystal structure of Type 1 human methionine aminopeptidase in complex with 2-(4-(5-chloro-6-methyl-2-(pyridin-2-yl)pyrimidin-4-yl)piperazin-1-yl)ethanol | | Descriptor: | 2-{4-[5-chloro-6-methyl-2-(pyridin-2-yl)pyrimidin-4-yl]piperazin-1-yl}ethanol, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Kishor, C, Arya, T. | | Deposit date: | 2012-12-28 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification, Biochemical and Structural Evaluation of Species-Specific Inhibitors against Type I Methionine Aminopeptidases
J.Med.Chem., 56, 2013
|
|
7ZQZ
 
 | |
7ZSW
 
 | |
3E6O
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C124355 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-[2-(4-AMINO-5,8-DIFLUORO-1,2-DIHYDROQUINAZOLIN-2-YL)ETHYL]-3-FURAMIDE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3ZEB
 
 | | A complex of GlpG with isocoumarin inhibitor covalently bonded to serine 201 and histidine 150 | | Descriptor: | 2-phenylethyl 2-(4-azanyl-2-methanoyl-phenyl)ethanoate, CHLORIDE ION, RHOMBOID PROTEASE GLPG, ... | | Authors: | Vinothkumar, K.R, voskya, O, Kuettler, E.V, Brouwer, A.J, Liskamp, R.M.J, Verhelst, S.H.L. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity-Based Probes for Rhomboid Proteases Discovered in a Mass Spectrometry-Based Assay.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4IBS
 
 | | Human p53 core domain with hot spot mutation R273H (form I) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Rozenberg, H, Eldar, A, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
2W04
 
 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with citrate in ATP binding site from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, CITRATE ANION | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
7ZLS
 
 | | co-crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 13 | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Ramachandran, S, Ciulli, A, Makukhin, N. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
3ZS5
 
 | | Structural basis for kinase selectivity of three clinical p38alpha inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MITOGEN-ACTIVATED PROTEIN KINASE 14, ... | | Authors: | Azevedo, R, van Zeeland, M, Raaijmakers, H.C.A, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
4IGK
 
 | |
2W91
 
 | | Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D. | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE D, PENTAETHYLENE GLYCOL | | Authors: | Abbott, D.W, Macauley, M.S, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Streptococcus Pneumoniae Endohexosaminidase D, Structural and Mechanistic Insight Into Substrate-Assisted Catalysis in Family 85 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
5DGD
 
 | |
4IHO
 
 | | Crystal structure of H-2Db Y159F in complex with chimeric gp100 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Uchtenhagen, H, Stahl, E, Achour, A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Proline substitution independently enhances H-2D(b) complex stabilization and TCR recognition of melanoma-associated peptides
Eur.J.Immunol., 43, 2013
|
|
