6ZVF
 
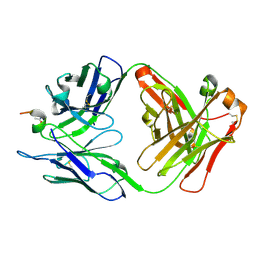 | |
6SRS
 
 | | Structure of the Fanconi anaemia core subcomplex | | 分子名称: | Fanconi anaemia protein FANCL, Unassigned secondary structure elements (central region, proposed FANCB-FAAP100), ... | | 著者 | Shakeel, S, Rajendra, E, Alcon, P, He, S, Scheres, S.H.W, Passmore, L.A. | | 登録日 | 2019-09-05 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structure of the Fanconi anaemia monoubiquitin ligase complex.
Nature, 575, 2019
|
|
6PJ1
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-4(AJ-74) | | 分子名称: | 1,2-ETHANEDIOL, 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2019-06-27 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
7RQE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MAI-tripeptidyl-tRNA analog ACCA-IAM, and chloramphenicol at 2.40A resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | 登録日 | 2021-08-06 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
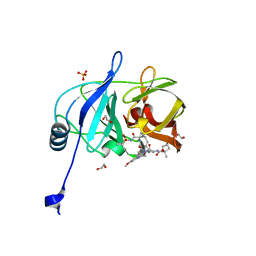
2AD9
 
 | | Solution structure of Polypyrimidine Tract Binding protein RBD1 complexed with CUCUCU RNA | | 分子名称: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | 著者 | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | 登録日 | 2005-07-20 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
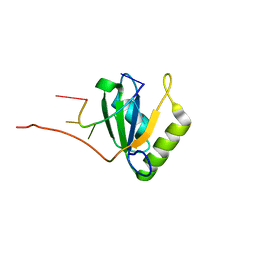
8TJO
 
 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 1 in complex with antibody fragment 1B2: Crosslinked Intra-State 1 | | 分子名称: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | 著者 | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | 登録日 | 2023-07-23 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 2024
|
|
7LW4
 
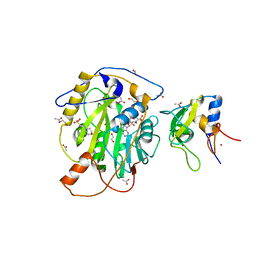 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of S-adenosyl-L-homocysteine (SAH) | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | 登録日 | 2021-02-27 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
6SWR
 
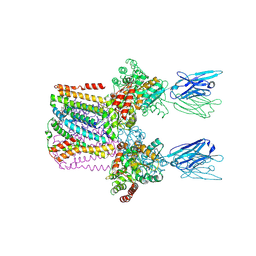 | | Crystal structure of the lysosomal potassium channel MtTMEM175 T38A mutant soaked with zinc | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Nanobody, Maltose/maltodextrin-binding periplasmic protein,Maltodextrin-binding protein,Maltose/maltodextrin-binding periplasmic protein, ... | | 著者 | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | 登録日 | 2019-09-23 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for ion selectivity in TMEM175 K + channels.
Elife, 9, 2020
|
|
6ESV
 
 | | Structure of the phosphate-bound form of AioX from Rhizobium sp. str. NT-26 | | 分子名称: | PHOSPHATE ION, Putative periplasmic phosphite-binding-like protein (Pbl) PtxB-like protein designated AioX | | 著者 | Djordjevic, S, Badilla, C, Cole, A, Santini, J. | | 登録日 | 2017-10-24 | | 公開日 | 2018-10-17 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A new family of periplasmic-binding proteins that sense arsenic oxyanions.
Sci Rep, 8, 2018
|
|
1FO5
 
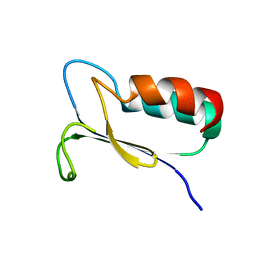 | | SOLUTION STRUCTURE OF REDUCED MJ0307 | | 分子名称: | THIOREDOXIN | | 著者 | Cave, J.W, Cho, H.S, Batchelder, A.M, Kim, R, Yokota, H, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2000-08-24 | | 公開日 | 2001-04-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution nuclear magnetic resonance structure of a protein disulfide oxidoreductase from Methanococcus jannaschii.
Protein Sci., 10, 2001
|
|
5M3M
 
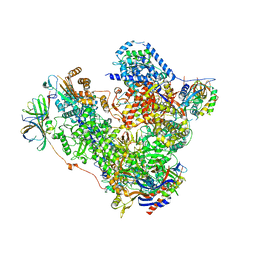 | | Free monomeric RNA polymerase I at 4.0A resolution | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Neyer, S, Kunz, M, Geiss, C, Hantsche, M, Hodirnau, V.-V, Seybert, A, Engel, C, Scheffer, M.P, Cramer, P, Frangakis, A.S. | | 登録日 | 2016-10-15 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of RNA polymerase I transcribing ribosomal DNA genes.
Nature, 540, 2016
|
|
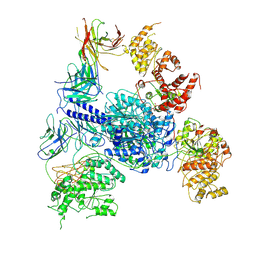
8G2W
 
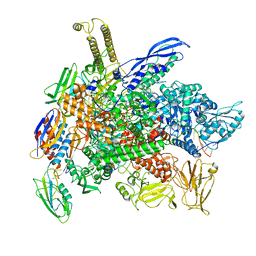 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-06 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7LVQ
 
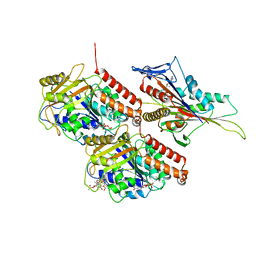 | | KIF14[391-743] - AMP-PNP closed state class in complex with a microtubule | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF14, ... | | 著者 | Benoit, M.P.M.H, Asenjo, A.B, Paydar, M, Dhakal, S, Kwok, B, Sosa, H. | | 登録日 | 2021-02-26 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14.
Nat Commun, 12, 2021
|
|
7V07
 
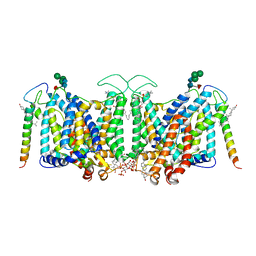 | | Band 3-I-TM local refinement from erythrocyte ankyrin-1 complex consensus reconstruction | | 分子名称: | Band 3 anion transport protein, CHOLESTEROL, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | 著者 | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | 登録日 | 2022-05-10 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5KBK
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), E110A Mutant | | 分子名称: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | 著者 | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | 登録日 | 2016-06-03 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.411 Å) | | 主引用文献 | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
7LOX
 
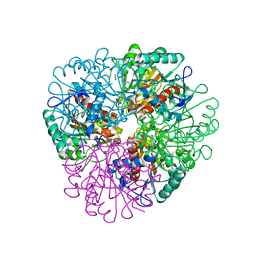 | | The structure of Agmatinase from E. Coli at 3.2 A displaying guanidine in the active site | | 分子名称: | Agmatinase, GUANIDINE, MANGANESE (II) ION | | 著者 | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | 登録日 | 2021-02-11 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
5LZS
 
 | | Structure of the mammalian ribosomal elongation complex with aminoacyl-tRNA, eEF1A, and didemnin B | | 分子名称: | (2~{S})-~{N}-[(2~{R})-1-[[(3~{S},6~{S},8~{S},12~{S},13~{R},16~{S},17~{R},20~{S},23~{S})-13-[(2~{S})-butan-2-yl]-20-[(4-methoxyphenyl)methyl]-6,17,21-trimethyl-3-(2-methylpropyl)-12-oxidanyl-2,5,7,10,15,19,22-heptakis(oxidanylidene)-8-propan-2-yl-9,18-dioxa-1,4,14,21-tetrazabicyclo[21.3.0]hexacosan-16-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-~{N}-methyl-1-[(2~{S})-2-oxidanylpropanoyl]pyrrolidine-2-carboxamide, 18S ribosomal RNA, 28S ribosomal RNA, ... | | 著者 | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2016-10-02 | | 公開日 | 2016-11-30 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5K0X
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | 分子名称: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | 著者 | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | 登録日 | 2016-05-17 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.231 Å) | | 主引用文献 | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
3JCB
 
 | | Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5 | | 分子名称: | Antibody 36D5 heavy chain, Antibody 36D5 light chain, Envelope glycoprotein gp120, ... | | 著者 | Hu, G, Liu, J, Roux, K, Taylor, K.A. | | 登録日 | 2015-11-25 | | 公開日 | 2017-05-10 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Structure of Simian Immunodeficiency Virus Envelope Spikes Bound with CD4 and Monoclonal Antibody 36D5.
J. Virol., 91, 2017
|
|
6SEB
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG | | 分子名称: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.272 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
5K1U
 
 | |
6XQ1
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | 分子名称: | 3-[3-({[3-(4-carboxyphenoxy)phenyl]methoxy}methyl)phenyl]-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | 著者 | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | 登録日 | 2020-07-09 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
1W6O
 
 | | X-RAY CRYSTAL STRUCTURE OF C2S HUMAN GALECTIN-1 COMPLEXED WITH LACTOSE | | 分子名称: | BETA-MERCAPTOETHANOL, GALECTIN-1, SULFATE ION, ... | | 著者 | Lopez-Lucendo, M.I.F, Gabius, H.J, Romero, A. | | 登録日 | 2004-08-20 | | 公開日 | 2004-10-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding
J.Mol.Biol., 343, 2004
|
|
8G4W
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-10 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6XC9
 
 | | Immune receptor complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Tresoldi, M.C.E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | 登録日 | 2020-06-08 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
