6XAT
 
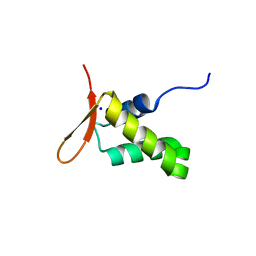 | | Crystal structure of the human FoxP4 DNA binding Domain | | Descriptor: | FOXP4 protein, SODIUM ION | | Authors: | VIllalobos, P, Castro-Fernandez, V, Medina, E, Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unraveling the folding and dimerization properties of the human FoxP subfamily of transcription factors.
Febs Lett., 597, 2023
|
|
6C8Z
 
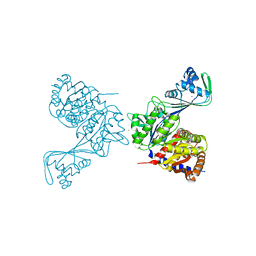 | | Last common ancestor of ADP-dependent phosphofructokinases from Methanosarcinales | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-dependent phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Castro-Fernandez, V, Gonzalez-Ordenes, F, Munoz, S, Fuentes, N, Leonardo, D, Fuentealba, M, Herrera-Morande, A, Maturana, P, Villalobos, P, Garratt, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ADP-Dependent Kinases From the Archaeal OrderMethanosarcinalesAdapt to Salt by a Non-canonical Evolutionarily Conserved Strategy.
Front Microbiol, 9, 2018
|
|
6XIO
 
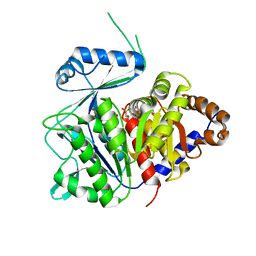 | | ADP-dependent kinase complex with fructose-6-phosphate and ADPbetaS | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, 6-O-phosphono-beta-D-fructofuranose, ADP-dependent phosphofructokinase, ... | | Authors: | Munoz, S, Gonzalez-Ordenes, F, Fuentes, N, Maturana, P, Herrera-Morande, A, Villalobos, P, Castro-Fernandez, V. | | Deposit date: | 2020-06-20 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of an ancestral ADP-dependent kinase with fructose-6P reveals key residues for binding, catalysis, and ligand-induced conformational changes.
J.Biol.Chem., 296, 2020
|
|
7LOX
 
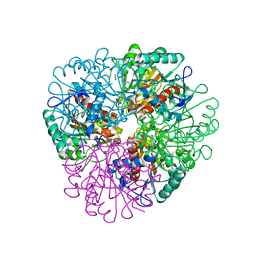 | | The structure of Agmatinase from E. Coli at 3.2 A displaying guanidine in the active site | | Descriptor: | Agmatinase, GUANIDINE, MANGANESE (II) ION | | Authors: | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | Deposit date: | 2021-02-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
7LOL
 
 | | The structure of Agmatinase from E. Coli at 1.8 A displaying urea and agmatine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGMATINE, Agmatinase, ... | | Authors: | Maturana, P, Figueroa, M, Gonzalez-Ordenes, F, Villalobos, P, Martinez-Oyanedel, J, Uribe, E.A, Castro-Fernandez, V. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity.
Int J Mol Sci, 22, 2021
|
|
