5AGS
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct 3-(Aminomethyl)-4-bromo-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 3-(AMINOMETHYL)-4-BROMO-7-ETHOXYBENZO[C][1,2]OXABOROL-1(3H)-OL-MODIFIED ADENOSINE, LEUCYL-TRNA SYNTHETASE, METHIONINE | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5GQT
 
 | |
6W4Q
 
 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
2DI3
 
 | |
3LQQ
 
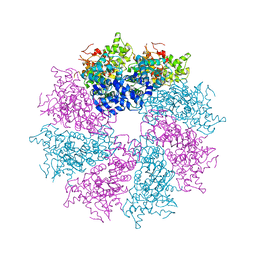 | | Structure of the CED-4 Apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N, Liu, Q. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.534 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
8WPG
 
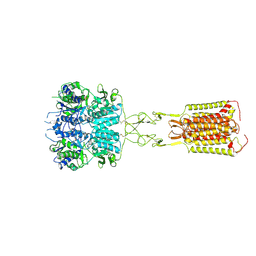 | | Human calcium-sensing receptor bound with cinacalcet in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPU
 
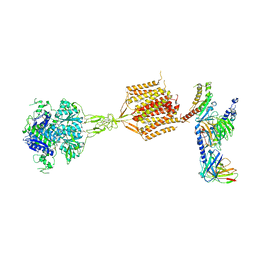 | | Human calcium-sensing receptor(CaSR) bound to cinacalcet in complex with Gq protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
4WRS
 
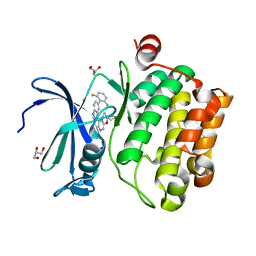 | |
3LQR
 
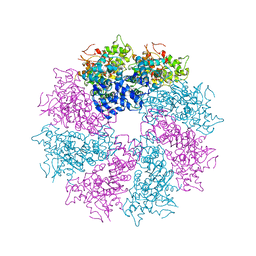 | | Structure of CED-4:CED-3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.896 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
6VC9
 
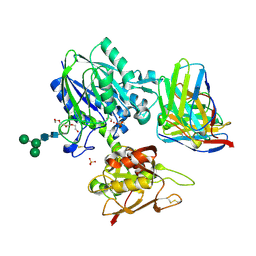 | | TB19 complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-nucleotidase, ecto (CD73), ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
6VCA
 
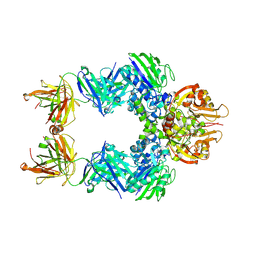 | | TB38 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
4YES
 
 | |
5GZE
 
 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
5GZG
 
 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
5GZC
 
 | | Crystal structure of Galectin-8 N-CRD with part of linker | | Descriptor: | GLYCEROL, Galectin-8, SODIUM ION | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
5GZD
 
 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
5GZF
 
 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
5JIE
 
 | |
2JXW
 
 | |
7LUB
 
 | |
7MBH
 
 | | Structure of Human Enolase 2 in complex with phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Gamma-enolase, ... | | Authors: | Leonard, P.G, Hicks, K.G, Rutter, J. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase.
Science, 379, 2023
|
|
7MSB
 
 | | Structure of EED bound to EEDi-4259 | | Descriptor: | (9aM,12aS)-12-{[(5-fluoro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,11,12a-tetraazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
