8F5P
 
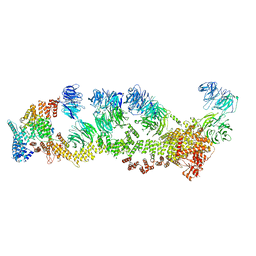 | | Structure of Leishmania tarentolae IFT-A (state 2) | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 122B, putative, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2022-11-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of IFT-A polymerization into trains for ciliary transport.
Cell, 185, 2022
|
|
8F5O
 
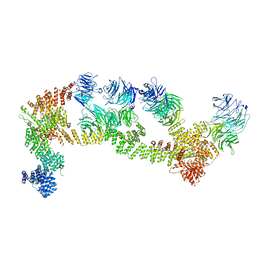 | | Structure of Leishmania tarentolae IFT-A (state 1) | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 122B, putative, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2022-11-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of IFT-A polymerization into trains for ciliary transport.
Cell, 185, 2022
|
|
7VS3
 
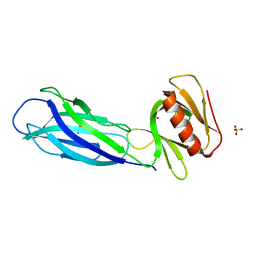 | | The crystal structure of rat calcium-dependent activator protein for secretion (CAPS) C2PH | | Descriptor: | Calcium-dependent secretion activator 1, SULFATE ION | | Authors: | Zhou, H, Wei, Z.Q, Zhang, L, Ren, Y.J, Ma, C. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The C 2 and PH domains of CAPS constitute an effective PI(4,5)P2-binding unit essential for Ca 2+ -regulated exocytosis.
Structure, 31, 2023
|
|
6L9J
 
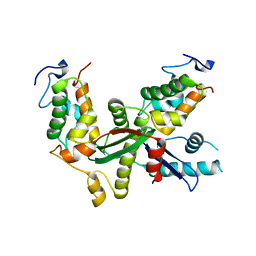 | | Structure of yeast Snf5 and Swi3 subcomplex | | Descriptor: | GLYCEROL, SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF complex subunit SWI3 | | Authors: | Long, J, Zhou, H. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Snf5 and Swi3 subcomplex formation is required for SWI/SNF complex function in yeast.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
6PWC
 
 | | A complex structure of arrestin-2 bound to neurotensin receptor 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Yin, W, Li, Z, Jin, M, Yin, Y.-L, de Waal, P.W, Pal, K, Gao, X, He, Y, Gao, J, Wang, X, Zhang, Y, Zhou, H, Melcher, K, Jiang, Y, Cong, Y, Zhou, X.E, Yu, X, Xu, H.E. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | A complex structure of arrestin-2 bound to a G protein-coupled receptor.
Cell Res., 29, 2019
|
|
1GMY
 
 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
6DXK
 
 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
7MD4
 
 | | Insulin receptor ectodomain dimer complexed with two IRPA-3 partial agonists | | Descriptor: | Insulin B chain, Insulin chain A, Isoform Short of Insulin receptor, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
7MD5
 
 | | Insulin receptor ectodomain dimer complexed with two IRPA-9 partial agonists | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
2V0G
 
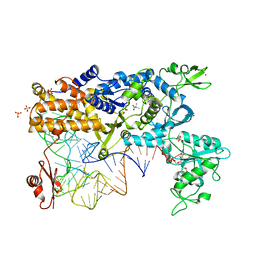 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A tRNA(leu) transcript with 5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1- BENZOXABOROLE (AN2690) forming an adduct to the ribose of adenosine- 76 in the enzyme editing site. | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, MERCURY (II) ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
2V0C
 
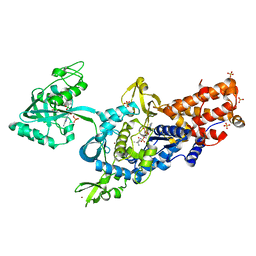 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A SULPHAMOYL ANALOGUE OF LEUCYL-ADENYLATE In the synthetic site and an adduct of AMP with 5-Fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690) in the editing site | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, SULFATE ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
1K0S
 
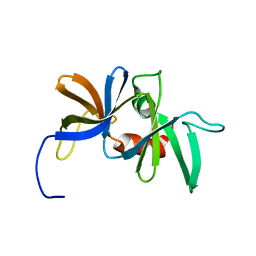 | | Solution structure of the chemotaxis protein CheW from the thermophilic organism Thermotoga maritima | | Descriptor: | CHEMOTAXIS PROTEIN CHEW | | Authors: | Griswold, I.J, Zhou, H, Swanson, R.V, Simon, M.I, Dahlquist, F.W. | | Deposit date: | 2001-09-20 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure and interactions of CheW from Thermotoga maritima.
Nat.Struct.Biol., 9, 2002
|
|
4REG
 
 | |
5GM2
 
 | | Crystal structure of methyltransferase TleD complexed with SAH and teleocidin A1 | | Descriptor: | (2S,5S)-9-[(3R)-3,7-dimethylocta-1,6-dien-3-yl]-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
5GM1
 
 | | Crystal structure of methyltransferase TleD complexed with SAH | | Descriptor: | O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
1NK1
 
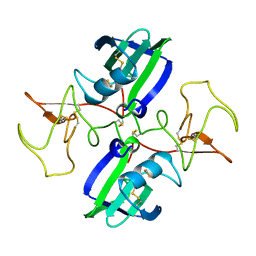 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
8YA4
 
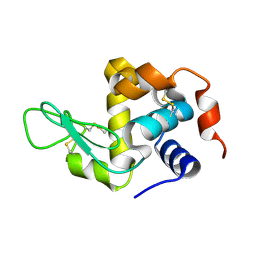 | |
8Y9V
 
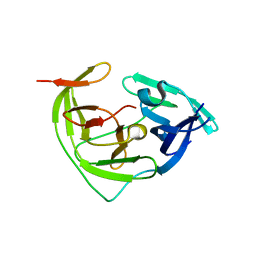 | | ZIKV NS2B/NS3 protease | | Descriptor: | DAR-LYS-ORN-ARG, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H, Wang, Q.S. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
8Y9W
 
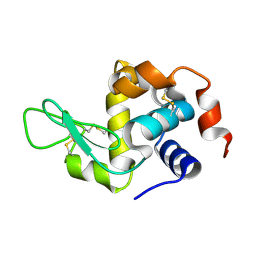 | |
8YA1
 
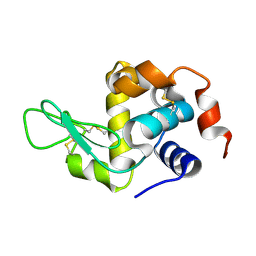 | | HEN EGG WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
8YA5
 
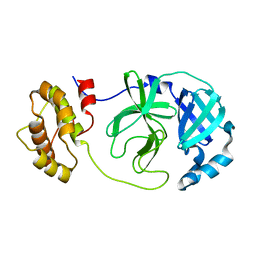 | |
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
8DK3
 
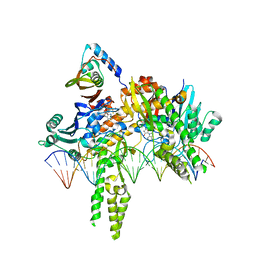 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetC ATPase domain bound to DNA and cWHD domain of JetA | | Descriptor: | DNA (26-MER), JetA, JetC, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
8DK2
 
 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetABC in an unclamped state trapped in ATP dependent dimeric form | | Descriptor: | DNA (26-MER), JetA, JetB, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
8DK1
 
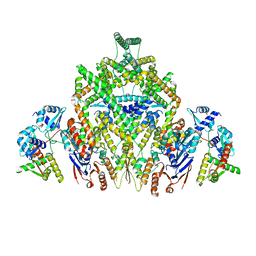 | | CryoEM structure of JetABC (head construct) from Pseudomonas aeruginosa PA14 | | Descriptor: | JetA, JetB, JetC | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
