5CD8
 
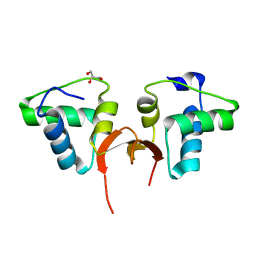 | | Crystal structure of the NTD of Drosophila Oskar protein | | Descriptor: | GLYCEROL, Maternal effect protein oskar | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CD9
 
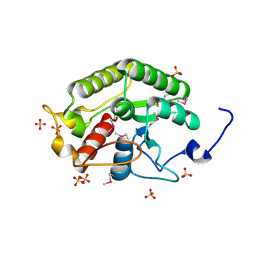 | | Crystal structure of the CTD of Drosophila Oskar protein | | Descriptor: | Maternal effect protein oskar, SULFATE ION | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
4JS9
 
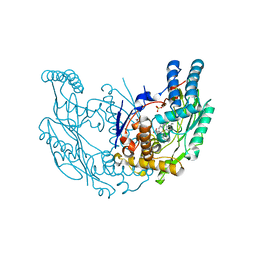 | | Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, Mesoheme, Nitric oxide synthase, ... | | Authors: | Hannibal, L, Page, R.C, Bolisetty, K, Yu, Z, Misra, S, Stuehr, D.J. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Kinetic and Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme
To be Published
|
|
5EX7
 
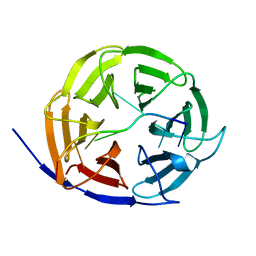 | | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA | | Descriptor: | Brain tumor protein, RNA (5'-R(P*UP*UP*UP*GP*UP*UP*GP*U)-3') | | Authors: | Wang, Y, Yu, Z, Wang, M, Liu, C.P, Yang, N, Xu, R.M. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA
To Be Published
|
|
5GRR
 
 | | Crystal structure of MCR-1 | | Descriptor: | GLYCEROL, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Ma, G, Zhu, Y, Yu, Z, Zhang, H. | | Deposit date: | 2016-08-12 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the catalytic domain of MCR-1
Sci Rep, 6, 2016
|
|
4PKM
 
 | | Crystal Structure of Bacillus thuringiensis Cry51Aa1 Protoxin at 1.65 Angstroms Resolution | | Descriptor: | Cry51Aa1, GLYCEROL, GLYCINE, ... | | Authors: | Xu, C, Chinte, U, Chen, L, Yao, Q, Zhou, D, Meng, Y, Li, L, Rose, J, Bi, L.J, Yu, Z, Sun, M, Wang, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Cry51Aa1: A potential novel insecticidal aerolysin-type beta-pore-forming toxin from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 462, 2015
|
|
4PVG
 
 | |
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7E9F
 
 | | Cryo-EM structure of the 2:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
7E9C
 
 | | Cryo-EM structure of the 1:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
6K9L
 
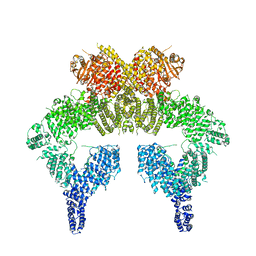 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
6NJP
 
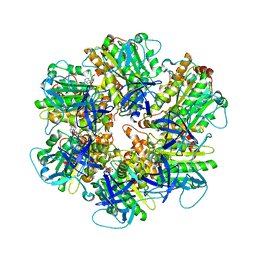 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
7Y8R
 
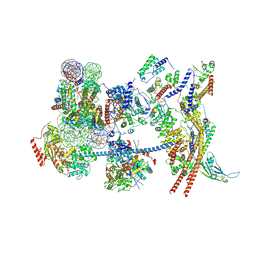 | | The nucleosome-bound human PBAF complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2, ... | | Authors: | Wang, L, Yu, J, Yu, Z, Wang, Q, He, S, Xu, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of nucleosome-bound human PBAF complex.
Nat Commun, 13, 2022
|
|
8HMB
 
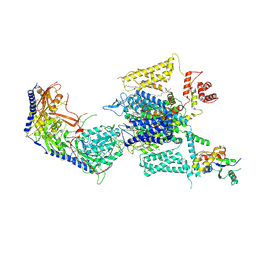 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | Descriptor: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
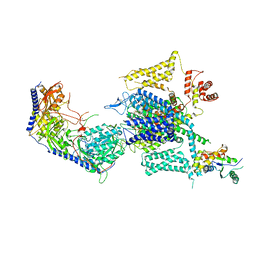 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HLP
 
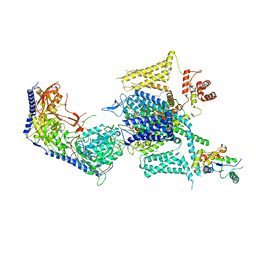 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
6B6H
 
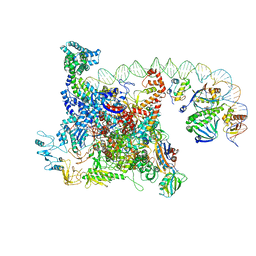 | | The cryo-EM structure of a bacterial class I transcription activation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
7DL2
 
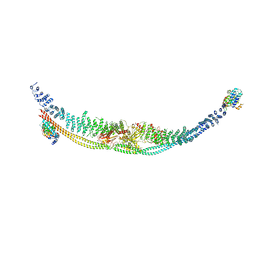 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
7DN3
 
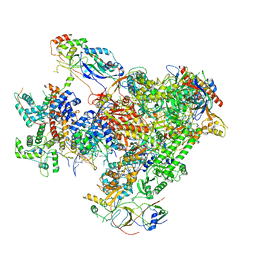 | | Structure of Human RNA Polymerase III elongation complex | | Descriptor: | DNA (5'-D(P*TP*CP*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*TP*CP*GP*GP*G)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
7DU2
 
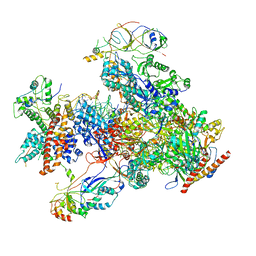 | | RNA polymerase III EC complex in post-translocation state | | Descriptor: | DNA (5'-D(P*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*T)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
7JR9
 
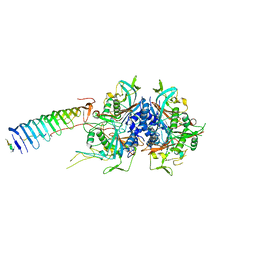 | | Chlamydomonas reinhardtii radial spoke minimal head complex | | Descriptor: | Flagellar radial spoke protein 10, Flagellar radial spoke protein 4, Flagellar radial spoke protein 6, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
