5XLT
 
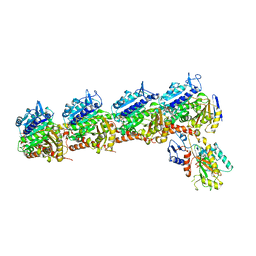 | | The crystal structure of tubulin in complex with 4'-demethylepipodophyllotoxin | | Descriptor: | (5S,5aR,8aR,9R)-9-(3,5-dimethoxy-4-oxidanyl-phenyl)-5-oxidanyl-5a,6,8a,9-tetrahydro-5H-[2]benzofuro[6,5-f][1,3]benzodioxol-8-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Structure of 4'-demethylepipodophyllotoxin in complex with tubulin provides a rationale for drug design
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6JMD
 
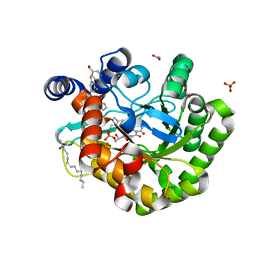 | | Crystal structure of human DHODH in complex with inhibitor 1223 | | Descriptor: | 3-[3,5-bis(fluoranyl)-4-[3-(hydroxymethyl)phenyl]phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
6JME
 
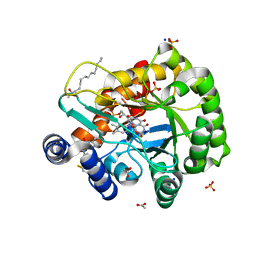 | | Crystal structure of human DHODH in complex with inhibitor 0946 | | Descriptor: | 3-[3,5-bis(fluoranyl)-4-(2-fluorophenyl)phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
7XRJ
 
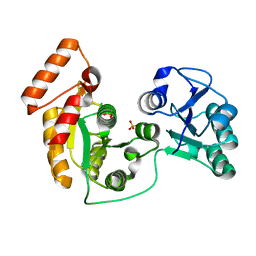 | | crystal structure of N-acetyltransferase DgcN-25328 | | Descriptor: | Putative NAD-dependent epimerase/dehydratase family protein, SULFATE ION | | Authors: | Zhang, Y.Z, Yu, Y, Cao, H.Y, Chen, X.L, Wang, P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel D-glutamate catabolic pathway in marine Proteobacteria and halophilic archaea.
Isme J, 17, 2023
|
|
6J3B
 
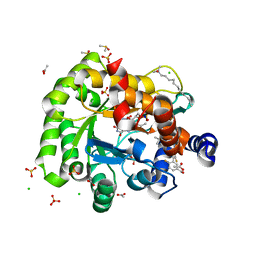 | | Crystal structure of human DHODH in complex with inhibitor 1289 | | Descriptor: | (6R)-1-[3,5-bis(fluoranyl)-4-[2-fluoranyl-5-(hydroxymethyl)phenyl]phenyl]-6-propan-2-yl-6,7-dihydro-5H-benzotriazol-4-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel series of human dihydroorotate dehydrogenase inhibitors discovered by in vitro screening: inhibition activity and crystallographic binding mode.
Febs Open Bio, 9, 2019
|
|
6J3C
 
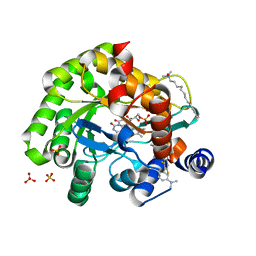 | | Crystal structure of human DHODH in complex with inhibitor 1291 | | Descriptor: | (6R)-1-[4-[3-(dimethylamino)phenyl]-3,5-bis(fluoranyl)phenyl]-6-propan-2-yl-6,7-dihydro-5H-benzotriazol-4-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A novel series of human dihydroorotate dehydrogenase inhibitors discovered by in vitro screening: inhibition activity and crystallographic binding mode.
Febs Open Bio, 9, 2019
|
|
6JBM
 
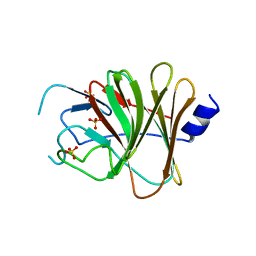 | | Crystal structure of the TRIM14 PRYSPRY domain | | Descriptor: | SULFATE ION, Tripartite motif-containing protein 14 | | Authors: | Yin, Y.X, Yu, Y, Liang, L. | | Deposit date: | 2019-01-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The TRIM14 PRYSPRY domain mediates protein interaction via its basic interface.
Febs Lett., 593, 2019
|
|
6K9V
 
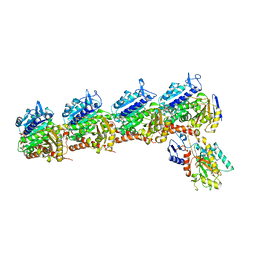 | | Crystal structure of tubulin in complex with inhibitor D64 | | Descriptor: | (5-methoxy-1H-indol-2-yl)-phenyl-methanone, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structural insights into the design of indole derivatives as tubulin polymerization inhibitors.
Febs Lett., 594, 2020
|
|
5YZ3
 
 | | Crystal structure of T2R-TTL-28 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2017-12-12 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | A Novel Microtubule Inhibitor Overcomes Multidrug Resistance in Tumors.
Cancer Res., 78, 2018
|
|
7CR6
 
 | | Synechocystis Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7CR8
 
 | | Synechocystis Cas1-Cas2-prespacerL complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
6KYQ
 
 | |
6KYR
 
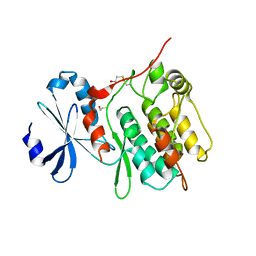 | |
7EI1
 
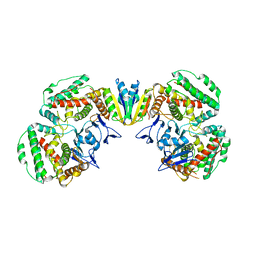 | | Structure of Pyrococcus furiosus Cas1Cas2 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-03-30 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A distinct structure of Cas1-Cas2 complex provides insights into the mechanism for the longer spacer acquisition in Pyrococcus furiosus.
Int.J.Biol.Macromol., 183, 2021
|
|
1AYG
 
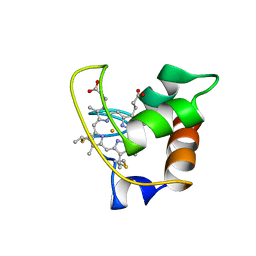 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
4V5M
 
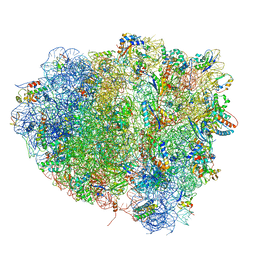 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
4V5N
 
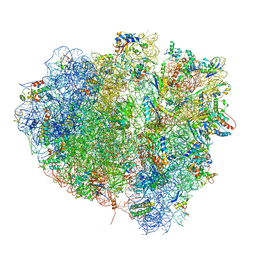 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
4WVR
 
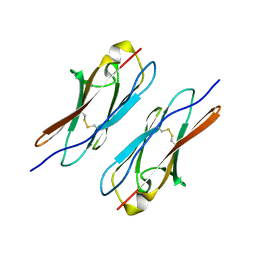 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
7MEY
 
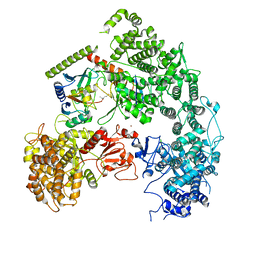 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | Descriptor: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
7MEX
 
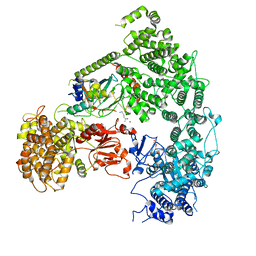 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
3DCG
 
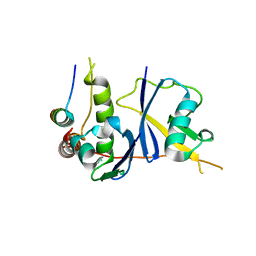 | | Crystal Structure of the HIV Vif BC-box in Complex with Human ElonginB and ElonginC | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Stanley, B.J, Ehrlich, E.S, Short, L, Yu, Y, Xiao, Z, Yu, X.-F, Xiong, Y. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly
J.Virol., 82, 2008
|
|
4EJQ
 
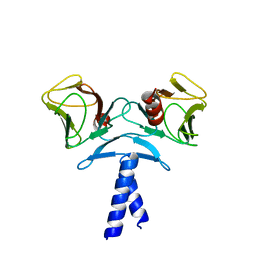 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
3N6N
 
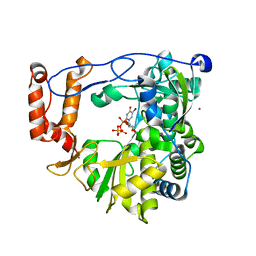 | | crystal structure of EV71 RdRp in complex with Br-UTP | | Descriptor: | 5-bromouridine 5'-(tetrahydrogen triphosphate), NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
3N6M
 
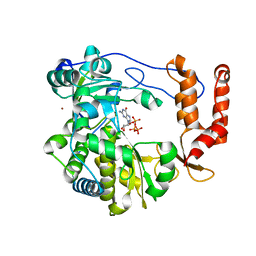 | | Crystal structure of EV71 RdRp in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
