2YT0
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2YS4
 
 | | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human | | Descriptor: | Hydrocephalus-inducing protein homolog | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human
To be Published
|
|
2YT1
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2YSO
 
 | | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog | | Descriptor: | ZINC ION, Zinc finger protein 95 homolog | | Authors: | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog
To be Published
|
|
2YSP
 
 | | Solution structure of the C2H2 type zinc finger (region 507-539) of human Zinc finger protein 224 | | Descriptor: | ZINC ION, Zinc finger protein 224 | | Authors: | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 507-539)of human Zinc finger protein 224
To be Published
|
|
2YT5
 
 | | Solution structure of the PHD domain of Metal-response element-binding transcription factor 2 | | Descriptor: | Metal-response element-binding transcription factor 2, ZINC ION | | Authors: | Masuda, K, Muto, Y, Isono, K, Watanabe, S, Harada, T, Kigawa, T, Koseki, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain of Metal-response element-binding transcription factor 2
To be Published
|
|
3KWQ
 
 | |
3KXB
 
 | |
7T8F
 
 | |
7T8E
 
 | | G93A mutant of human SOD1 in P21 space group | | Descriptor: | ACETATE ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Amporndanai, K, Hasnain, S.S. | | Deposit date: | 2021-12-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ebselen analogues delay disease onset and its course in fALS by on-target SOD-1 engagement.
Sci Rep, 14, 2024
|
|
7T8H
 
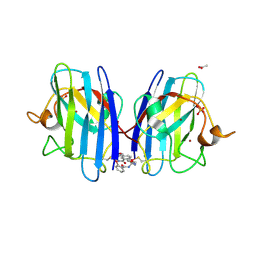 | |
7T8G
 
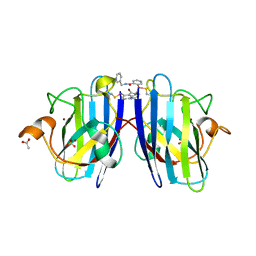 | |
3KC6
 
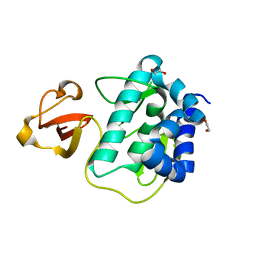 | |
3KHW
 
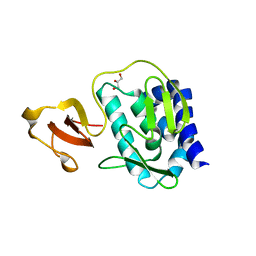 | |
3L56
 
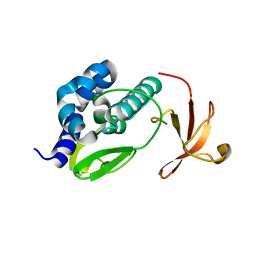 | | Crystal structure of the large c-terminal domain of polymerase basic protein 2 from influenza virus a/viet nam/1203/2004 (h5n1) | | Descriptor: | Polymerase PB2 | | Authors: | Staker, B.L, Edwards, T, Eric, S, Raymond, A, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-12-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biological and structural characterization of a host-adapting amino acid in influenza virus.
Plos Pathog., 6, 2010
|
|
2KUP
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | 19-residue peptide from ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
3MBC
 
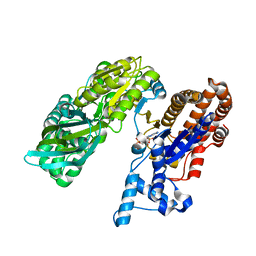 | | Crystal structure of monomeric isocitrate dehydrogenase from Corynebacterium glutamicum in complex with NADP | | Descriptor: | Isocitrate dehydrogenase [NADP], MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sidhu, N.S, Aich, S, Sheldrick, G.M, Delbaere, L.T.J. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly NADP+-specific isocitrate dehydrogenase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
8YHR
 
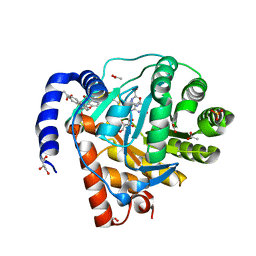 | | DHODH in complex with furocoumavirin | | Descriptor: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | Deposit date: | 2024-02-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
6R76
 
 | |
6R77
 
 | |
7FBR
 
 | |
7FBV
 
 | |
6K9Y
 
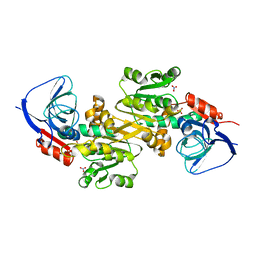 | | Crystal structure of human VAT-1 | | Descriptor: | NITRATE ION, Synaptic vesicle membrane protein VAT-1 homolog | | Authors: | Watanabe, Y, Endo, T. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for interorganelle phospholipid transport mediated by VAT-1.
J.Biol.Chem., 295, 2020
|
|
7EXZ
 
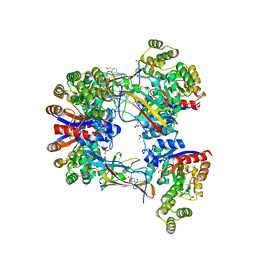 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
 | | DfgA-DfgB complex apo 2.4 angstrom | | Descriptor: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
