7TQS
 
 | |
4NCO
 
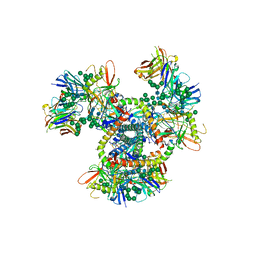 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP gp120, BG505 SOSIP gp41, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Lyumkis, D, Ward, A.B, Wilson, I.A. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Crystal structure of a soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
4NHG
 
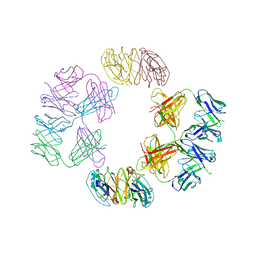 | | Crystal Structure of 2G12 IgG Dimer | | Descriptor: | 2G12 IgG dimer heavy chain, 2G12 IgG dimer light chain, Hepatitis B virus receptor binding protein | | Authors: | Wu, Y, West Jr, A.P, Kim, H.J, Thornton, M.E, Ward, A.B, Bjorkman, P.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12.
Cell Rep, 5, 2013
|
|
4NHH
 
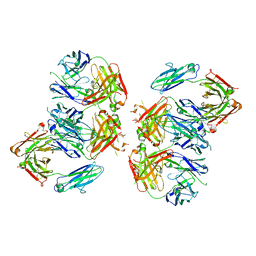 | | Structure of 2G12 IgG Dimer | | Descriptor: | 2G12 IgG dimer heavy chain, 2G12 IgG dimer light chain, Hepatitis B virus receptor binding protein | | Authors: | Wu, Y, West Jr, A.P, Kim, H.J, Thornton, M.E, Ward, A.B, Bjorkman, P.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12.
Cell Rep, 5, 2013
|
|
8EQN
 
 | |
8EJE
 
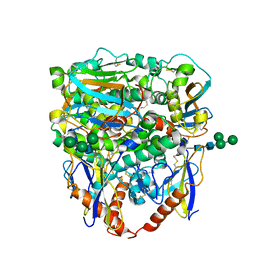 | |
8EJF
 
 | | Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies
Cell Rep, 42, 2023
|
|
8EJG
 
 | |
8EJD
 
 | | Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies
Cell Rep, 42, 2023
|
|
8EJI
 
 | |
8EJH
 
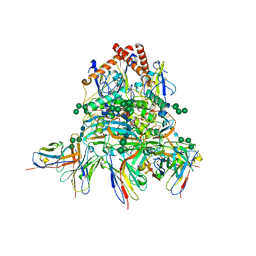 | |
8EJJ
 
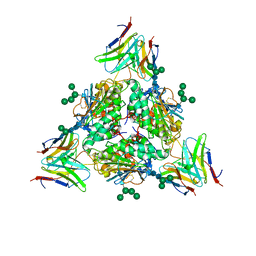 | | Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies
Cell Rep, 42, 2023
|
|
8DYT
 
 | | Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide | | Descriptor: | 227 Fab heavy chain, 227 Fab light chain, Circumsporozoite protein | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Affinity-matured homotypic interactions induce spectrum of PfCSP structures that influence protection from malaria infection.
Nat Commun, 14, 2023
|
|
8E6K
 
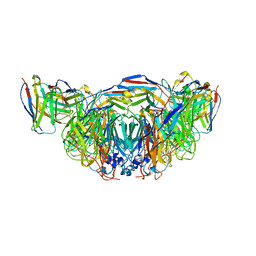 | | 2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2H08 fragment antigen binding heavy chain, 2H08 fragment antigen binding light chain, ... | | Authors: | Turner, H.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8DYX
 
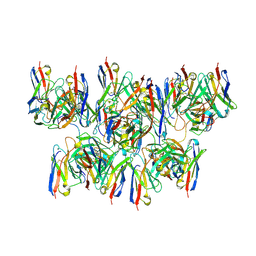 | |
8DZ4
 
 | |
8E6J
 
 | |
8DZ3
 
 | |
8DYY
 
 | |
8DZ5
 
 | |
8DYW
 
 | |
6OPO
 
 | | Symmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with DDM | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6OPQ
 
 | | CD4- and 17-bound HIV-1 Env B41 SOSIP frozen with LMNG | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6OPP
 
 | | Asymmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with DDM | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6OPN
 
 | | CD4- and 17-bound HIV-1 Env B41 SOSIP in complex with small molecule GO35 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
