4YQ2
 
 | |
4YQ9
 
 | |
4YQK
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 4-amino-N-(piperidin-4-yl)-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
4YQT
 
 | |
5COY
 
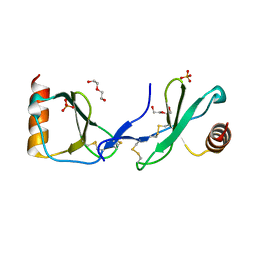 | | Crystal structure of CC chemokine 5 (CCL5) | | Descriptor: | C-C motif chemokine 5, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-07-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CMD
 
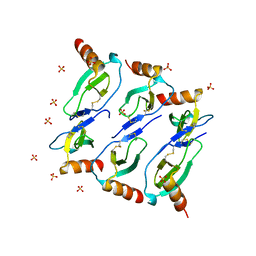 | |
5CJO
 
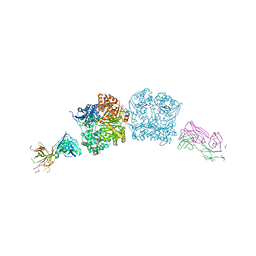 | |
5D9F
 
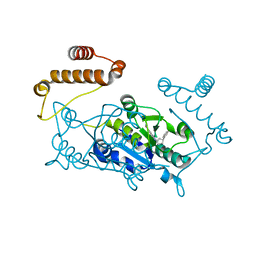 | |
5D65
 
 | | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-C motif chemokine 3, CHLORIDE ION, ... | | Authors: | Liang, W.G, Hwang, D.Y, Zulueta, M.M, Hung, S.C, Tang, W. | | Deposit date: | 2015-08-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DNF
 
 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RQC
 
 | | Crystals of peptide deformylase from Plasmodium falciparum with ten subunits per asymmetric unit reveal critical characteristics of the active site for drug design | | Descriptor: | COBALT (II) ION, formylmethionine deformylase | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G. | | Deposit date: | 2003-12-04 | | Release date: | 2004-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase
Protein Sci., 13, 2004
|
|
1TV6
 
 | | HIV-1 Reverse Transcriptase Complexed with CP-94,707 | | Descriptor: | 3-[4-(2-METHYL-IMIDAZO[4,5-C]PYRIDIN-1-YL)BENZYL]-3H-BENZOTHIAZOL-2-ONE, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit | | Authors: | Pata, J.D, Stirtan, W.G, Goldstein, S.W, Steitz, T.A. | | Deposit date: | 2004-06-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant RTs resistant to other non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VCB
 
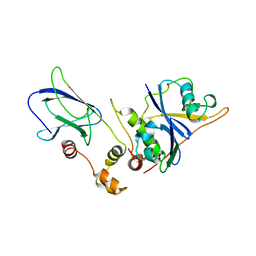 | | THE VHL-ELONGINC-ELONGINB STRUCTURE | | Descriptor: | PROTEIN (ELONGIN B), PROTEIN (ELONGIN C), PROTEIN (VHL) | | Authors: | Stebbins, C.E, Kaelin, W.G, Pavletich, N.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function.
Science, 284, 1999
|
|
4HSV
 
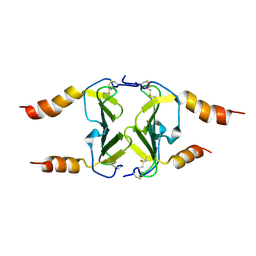 | | Crystal Structure of CXCL4L1 | | Descriptor: | Platelet factor 4 variant | | Authors: | Kuo, J.H, Liu, J.S, Wu, W.G, Sue, S.C. | | Deposit date: | 2012-10-30 | | Release date: | 2013-04-10 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Alternative C-terminal helix orientation alters chemokine function: structure of the anti-angiogenic chemokine, CXCL4L1
J.Biol.Chem., 288, 2013
|
|
2PMW
 
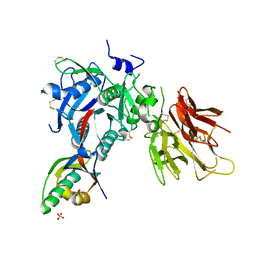 | | The Crystal Structure of Proprotein convertase subtilisin kexin type 9 (PCSK9) | | Descriptor: | Proprotein convertase subtilisin/kexin type 9, SULFATE ION | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of PCSK9: A Regulator of Plasma LDL-Cholesterol.
Structure, 15, 2007
|
|
4IFH
 
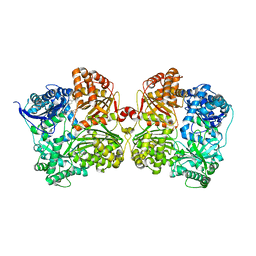 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 | | Descriptor: | Insulin-degrading enzyme, N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)-4-methylbenzamide, ZINC ION | | Authors: | Liang, W.G, Guo, Q, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4IOF
 
 | | Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) | | Descriptor: | Fab-bound IDE, heavy chain, light chain, ... | | Authors: | McCord, L.A, Liang, W.G, Hoey, R, Dowdell, E, Koide, A, Koide, S, Tang, W.J. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | Conformational states and recognition of amyloidogenic peptides of human insulin-degrading enzyme.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2QUS
 
 | |
2QUW
 
 | |
4K7A
 
 | |
4KSR
 
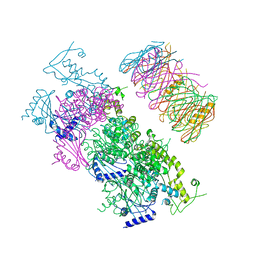 | | Crystal Structure of the Vibrio cholerae ATPase GspE Hexamer | | Descriptor: | Type II secretion system protein E, Hemolysin-coregulated protein | | Authors: | Hol, W.G, Turley, S, Lu, C.Y, Park, Y.J, Marionni, S.T, Lee, K, Patrick, M, Sandkvist, M, Bush, M, Shah, R. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Hexamers of the Type II Secretion ATPase GspE from Vibrio cholerae with Increased ATPase Activity.
Structure, 21, 2013
|
|
4KGO
 
 | |
4KSS
 
 | | Crystal Structure of Vibrio cholerae ATPase GspsE Hexamer | | Descriptor: | Type II secretion system protein E, hemolysin-coregulated protein | | Authors: | Hol, W.G, Turley, S, Lu, C.Y, Park, Y.J. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (7.58 Å) | | Cite: | Hexamers of the Type II Secretion ATPase GspE from Vibrio cholerae with Increased ATPase Activity.
Structure, 21, 2013
|
|
4KGH
 
 | |
4K59
 
 | |
