3FKU
 
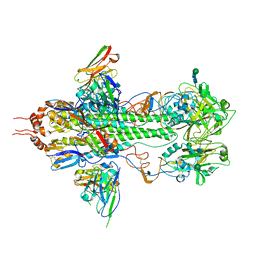 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Neutralizing antibody F10, ... | | Authors: | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2GHW
 
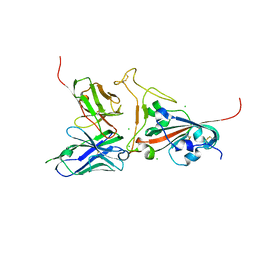 | | Crystal structure of SARS spike protein receptor binding domain in complex with a neutralizing antibody, 80R | | Descriptor: | CHLORIDE ION, Spike glycoprotein, anti-sars scFv antibody, ... | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2GHV
 
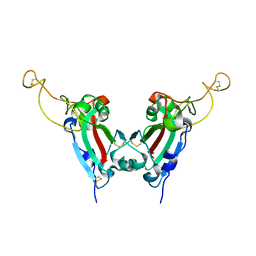 | | Crystal structure of SARS spike protein receptor binding domain | | Descriptor: | Spike glycoprotein | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
5YAX
 
 | | Crystal structure of a human neutralizing antibody bound to a HBV preS1 peptide | | Descriptor: | Large envelope protein, SODIUM ION, scFv1 antibody | | Authors: | Liu, X, Zheng, S, Ye, K, Sui, J. | | Deposit date: | 2017-09-02 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A potent human neutralizing antibody Fc-dependently reduces established HBV infections
Elife, 6, 2017
|
|
7WN8
 
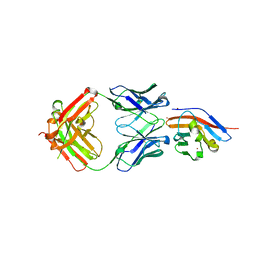 | | Crystal structure of antibody (BC31M5) binds to CD47 | | Descriptor: | BC31M5 Fab Heavy chain, BC31M5 Fab Light chain, Leukocyte surface antigen CD47, ... | | Authors: | Li, Y, Wang, W, Sui, J, Zhang, S. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A pH-dependent anti-CD47 antibody that selectively targets solid tumors and improves therapeutic efficacy and safety.
J Hematol Oncol, 16, 2023
|
|
6KTR
 
 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
7DF1
 
 | | Crystal structure of human CD98 heavy chain extracellular domain in complex with S1-F4 scFv | | Descriptor: | 4F2 cell-surface antigen heavy chain, IGL c2062_light_IGKV4-1_IGKJ5, S1-F4 VH | | Authors: | Liu, X, Ding, J, Sui, J, Tian, X. | | Deposit date: | 2020-11-06 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | An anti-CD98 antibody displaying pH-dependent Fc-mediated tumour-specific activity against multiple cancers in CD98-humanized mice.
Nat Biomed Eng, 2022
|
|
5ZV2
 
 | | FGFR-1 in complex with ligand lenvatinib | | Descriptor: | 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, Fibroblast growth factor receptor 1 | | Authors: | Matsuki, M, Hoshi, T, Yamamoto, Y, Ikemori-Kawada, M, Minoshima, Y, Funahashi, Y, Matsui, J. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models.
Cancer Med, 7, 2018
|
|
3WZE
 
 | | KDR in complex with ligand sorafenib | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, ACETATE ION, ... | | Authors: | Okamoto, K, Ikemori_Kawada, M, Inoue, A, Matsui, J. | | Deposit date: | 2014-09-24 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization.
ACS MED.CHEM.LETT., 6, 2015
|
|
3WZD
 
 | | KDR in complex with ligand lenvatinib | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, ... | | Authors: | Okamoto, K, Ikemori_Kawada, M, Inoue, A, Matsui, J. | | Deposit date: | 2014-09-24 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization.
ACS MED.CHEM.LETT., 6, 2015
|
|
7X8X
 
 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
7VQO
 
 | |
7DDE
 
 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
5X4N
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N4-phenylpyrimidine-2,4-diamine, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | B-cell lymphoma 6 protein, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4M
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | Descriptor: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4Q
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(pyridin-3-ylmethylamino)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 6 | | Descriptor: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
1IWP
 
 | | Glycerol Dehydratase-cyanocobalamin Complex of Klebsiella pneumoniae | | Descriptor: | COBALAMIN, Glycerol Dehydratase Alpha subunit, Glycerol Dehydratase Beta subunit, ... | | Authors: | Yamanishi, M, Yunoki, M, Tobimatsu, T, Toraya, T. | | Deposit date: | 2002-05-28 | | Release date: | 2002-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol.
Eur.J.Biochem., 269, 2002
|
|
