1SRK
 
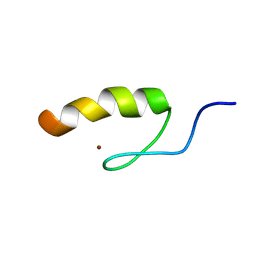 | | Solution structure of the third zinc finger domain of FOG-1 | | Descriptor: | ZINC ION, Zinc finger protein ZFPM1 | | Authors: | Simpson, R.J.Y, Lee, S.H.Y, Bartle, N, Matthews, J.M, Mackay, J.P, Crossley, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Classic Zinc Finger from Friend of GATA Mediates an Interaction with the Coiled-coil of Transforming Acidic Coiled-coil 3.
J.Biol.Chem., 279, 2004
|
|
5WUV
 
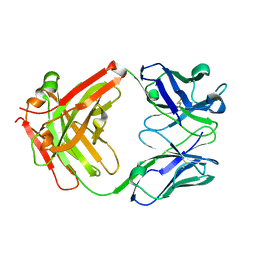 | | Crystal structure of Certolizumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S, Lee, J.U, Son, J.Y, Shin, W, Yoo, K.Y. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
6A6P
 
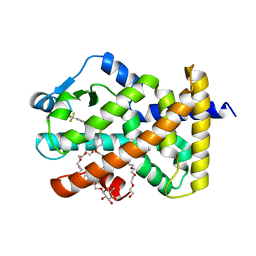 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd)LBD in Complex with DN003316 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside, ... | | Authors: | Chin, J.W, Cho, S.J, Song, J.Y, Ha, J.H. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd)LBD in Complex with DN003316
To Be Published
|
|
7CM0
 
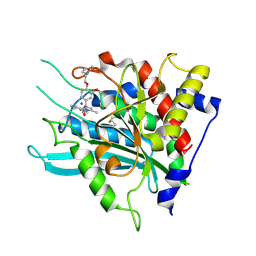 | | Crystal structure of a glutaminyl cyclase in complex with NHV-1009 | | Descriptor: | 1-(cyclopentylmethyl)-1-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]-3-[3-(5-methylimidazol-1-yl)propyl]urea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Lee, J.W, Song, J.Y, Jang, T.H, Ha, J.H. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of highly potent human glutaminyl cyclase (QC) inhibitors as anti-Alzheimer's agents by the combination of pharmacophore-based and structure-based design.
Eur.J.Med.Chem., 226, 2021
|
|
1Y0J
 
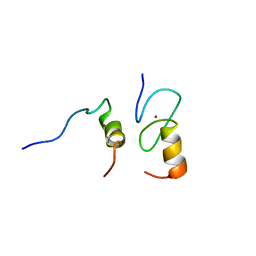 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1P7A
 
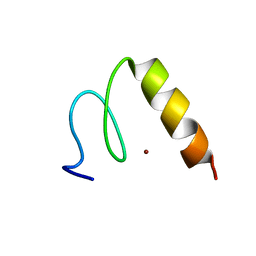 | | Solution Structure of the Third Zinc Finger from BKLF | | Descriptor: | Kruppel-like factor 3, ZINC ION | | Authors: | Simpson, R.J.Y, Cram, E.D, Czolij, R, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions.
J.Biol.Chem., 278, 2003
|
|
4Y7I
 
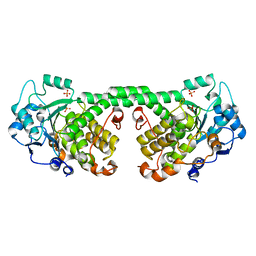 | | Crystal Structure of MTMR8 | | Descriptor: | Myotubularin-related protein 8, PHOSPHATE ION | | Authors: | Yoo, K, Lee, J, Son, J, Shin, W, Im, D, Heo, Y.S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the catalytic phosphatase domain of MTMR8: implications for dimerization, membrane association and reversible oxidation.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5GGS
 
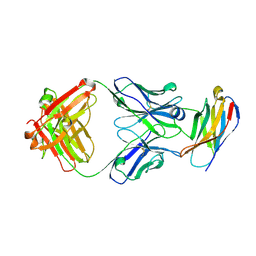 | | PD-1 in complex with pembrolizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGV
 
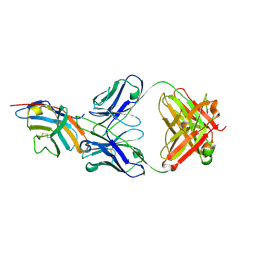 | | CTLA-4 in complex with tremelimumab Fab | | Descriptor: | Cytotoxic T-lymphocyte protein 4, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGT
 
 | | PD-L1 in complex with BMS-936559 Fab | | Descriptor: | IGK@ protein, IgG H chain, Programmed cell death 1 ligand 1 | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGR
 
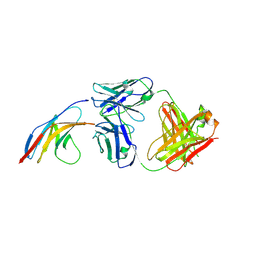 | | PD-1 in complex with nivolumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGQ
 
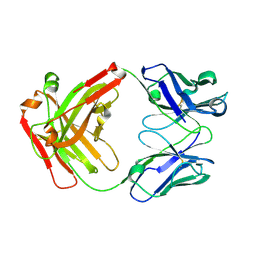 | | Crystal structure of Nivolumab Fab fragment | | Descriptor: | nivolumab heavy chain, nivolumab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGU
 
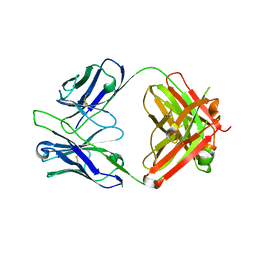 | | Crystal structure of tremelimumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5WUX
 
 | | TNFalpha-certolizumab Fab | | Descriptor: | Tumor necrosis factor alpha, heavy, light | | Authors: | Heo, Y.S, Lee, J.U. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
5Y9J
 
 | | BAFF in complex with belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belibumab light chain, belimumab heavy chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
5Y9K
 
 | | Structure of the belimumab Fab fragment | | Descriptor: | belimumab heavy chain, belimumab light chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
6ESL
 
 | | Crystal structure of the Legionella pneumoppila LapA | | Descriptor: | Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Richardson, K, Garnett, J.A. | | Deposit date: | 2017-10-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Type II Secretion-Dependent Aminopeptidase LapA and Acyltransferase PlaC Are Redundant for Nutrient Acquisition duringLegionella pneumophilaIntracellular Infection of Amoebas.
MBio, 9, 2018
|
|
6XTT
 
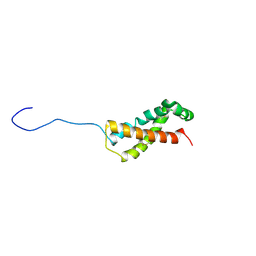 | |
6SJT
 
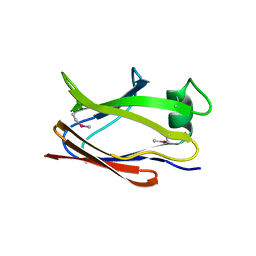 | |
6SKW
 
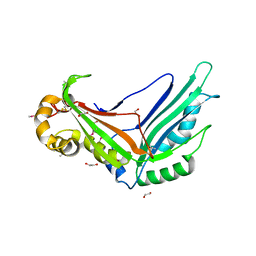 | | Crystal structure of the Legionella pneumophila type II secretion system substrate NttE | | Descriptor: | 1,2-ETHANEDIOL, NttE | | Authors: | Portlock, T.J, Rehman, S, Garnett, J.A. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Dynamics and Cellular Insight Into Novel Substrates of theLegionella pneumophilaType II Secretion System.
Front Mol Biosci, 7, 2020
|
|
7Z5H
 
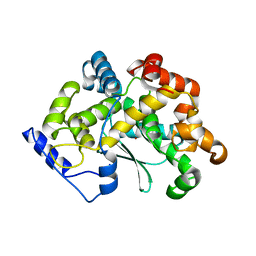 | | human Zn MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like, ZINC ION | | Authors: | Bak, J, Adamopoulos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z5G
 
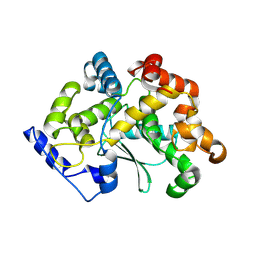 | | human apo MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like | | Authors: | Bak, J, Adamoupolos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z6S
 
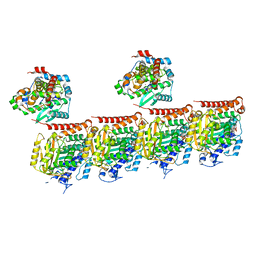 | | MATCAP bound to a human 14 protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bak, J, Perrakis, A. | | Deposit date: | 2022-03-14 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7COE
 
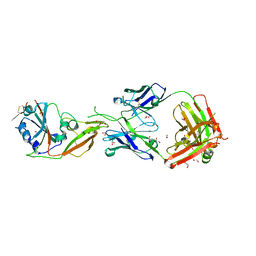 | | Crystal structure of Receptor binding domain of MERS-CoV and KNIH90-F1 Fab complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Lee, J.Y, Song, J.Y, Lee, H.S, Hong, E, Jang, T.H. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a novel antibody against the spike protein inhibits Middle East respiratory syndrome coronavirus infections.
Sci Rep, 12, 2022
|
|
2WKD
 
 | |
