3K7V
 
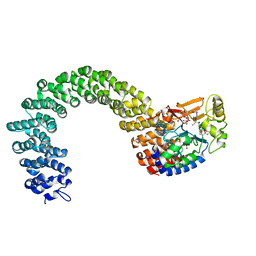 | | Protein phosphatase 2A core complex bound to dinophysistoxin-1 | | 分子名称: | (2R)-3-[(2S,5R,6R,8S)-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-6'-{(1S,3S)-3-[(2S,3R,6R,11R)-3,11-dimethyl-1,7-dioxaspiro[5.5]undec-2-yl]-1-hydroxybutyl}-8'-hydroxy-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]pyran]-5-yl]-1-methylprop-2-en-1-yl}-5-hydroxy-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-hydroxy-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | 著者 | Jeffrey, P.D, Huhn, J, Shi, Y. | | 登録日 | 2009-10-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
3JCM
 
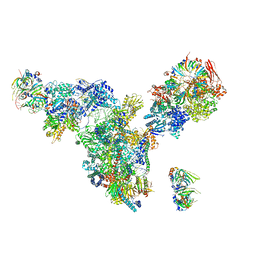 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | 著者 | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | 登録日 | 2015-12-23 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
4O6Y
 
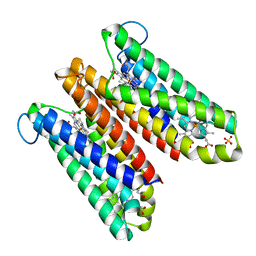 | | Crystal Structure of Cytochrome b561 | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, SULFATE ION | | 著者 | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | 登録日 | 2013-12-24 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O7G
 
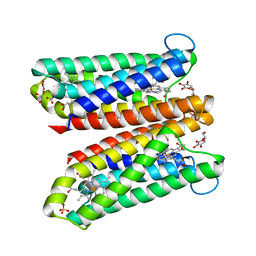 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 40 minutes | | 分子名称: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | 著者 | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | 登録日 | 2013-12-24 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.211 Å) | | 主引用文献 | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3KV5
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV9
 
 | | Structure of KIAA1718 Jumonji domain | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV4
 
 | | Structure of PHF8 in complex with histone H3 | | 分子名称: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVB
 
 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | 分子名称: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L1L
 
 | |
3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LRB
 
 | |
4Q9V
 
 | | Crystal structure of TIPE3 | | 分子名称: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | 著者 | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | 登録日 | 2014-05-02 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
3YGS
 
 | | APAF-1 CARD IN COMPLEX WITH PRODOMAIN OF PROCASPASE-9 | | 分子名称: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1, PROCASPASE 9 | | 著者 | Qin, H, Srinivasula, S, Wu, G, Fernandes-Alnemri, T, Alnemri, E, Shi, Y. | | 登録日 | 1999-05-08 | | 公開日 | 2000-04-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1.
Nature, 399, 1999
|
|
3LRC
 
 | |
4R7A
 
 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | 分子名称: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | 著者 | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | 登録日 | 2014-08-27 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
4RHW
 
 | | Crystal structure of Apaf-1 CARD and caspase-9 CARD complex | | 分子名称: | Apoptotic protease-activating factor 1, CHLORIDE ION, Caspase-9, ... | | 著者 | Hu, Q, Wu, D, Yan, C, Shi, Y. | | 登録日 | 2014-10-03 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
5WSG
 
 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | 分子名称: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | 著者 | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-12-07 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
6KLH
 
 | | Dimeric structure of Machupo virus polymerase bound to vRNA promoter | | 分子名称: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | 著者 | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | 登録日 | 2019-07-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
8GVB
 
 | | The complex between public TCR TD08 and HLA-A24 bound to HIV-1 Nef138-8 peptide | | 分子名称: | 8-mer peptide from Protein Nef, Beta-2-microglobulin, MHC class I antigen, ... | | 著者 | Gao, G.F, Shi, Y, Ma, K, Chai, Y, Guan, J, Qi, J, Tan, S, Dong, T, Iwamoto, A, Kawana-Tachikawa, A. | | 登録日 | 2022-09-14 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Molecular Basis for the Recognition of HIV Nef138-8 Epitope by a Pair of Human Public T Cell Receptors.
J Immunol., 209, 2022
|
|
6KLD
 
 | | Structure of apo Machupo virus polymerase | | 分子名称: | MANGANESE (II) ION, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | 登録日 | 2019-07-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6KLE
 
 | | Monomeric structure of Machupo virus polymerase bound to vRNA promoter | | 分子名称: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | 著者 | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | 登録日 | 2019-07-30 | | 公開日 | 2020-03-18 | | 最終更新日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
3NSS
 
 | | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active sites | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | 著者 | Li, Q, Qi, J.X, Zhang, W, Vavricka, C.J, Shi, Y, Gao, G.F. | | 登録日 | 2010-07-02 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site
Nat.Struct.Mol.Biol., 17, 2010
|
|
6KL9
 
 | | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form A complex) | | 分子名称: | AcrVA4, LbCas12a, MAGNESIUM ION, ... | | 著者 | Peng, R, Li, Z, Xu, Y, He, S, Peng, Q, Shi, Y, Gao, G.F. | | 登録日 | 2019-07-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KLB
 
 | | Structure of LbCas12a-crRNA complex bound to AcrVA4 (form B complex) | | 分子名称: | AcrVA4, LbCas12a, MAGNESIUM ION, ... | | 著者 | Peng, R, Li, Z, Xu, Y, He, S, Peng, Q, Shi, Y, Gao, G.F. | | 登録日 | 2019-07-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into multistage inhibition of CRISPR-Cas12a by AcrVA4.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
