1ZH1
 
 | |
1A1R
 
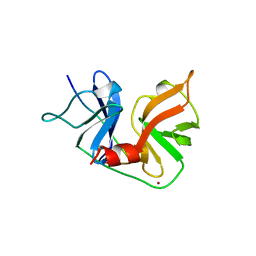 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
3KQH
 
 | |
3KQL
 
 | |
3KQK
 
 | |
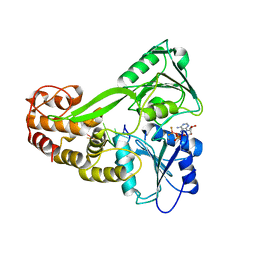
3KQN
 
 | |
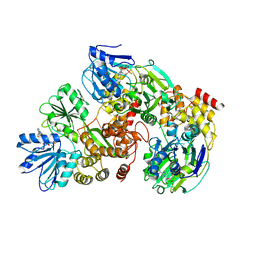
3KQU
 
 | |
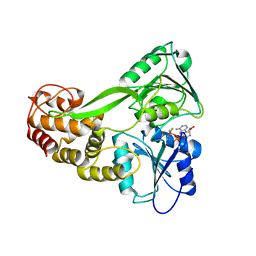
5E4F
 
 | |
4J1V
 
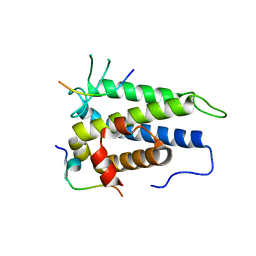 | | Functional and structural studies of MOBKL1B, a Salvador/Warts/Hippo tumor suppressor pathway, in HCV replication | | Descriptor: | MOB kinase activator 1A, NS5A domain II peptide, ZINC ION | | Authors: | Chung, H.-Y, Gu, M, Rice, C.M. | | Deposit date: | 2013-02-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Seed Sequence-Matched Controls Reveal Limitations of Small Interfering RNA Knockdown in Functional and Structural Studies of Hepatitis C Virus NS5A-MOBKL1B Interaction.
J.Virol., 88, 2014
|
|
2HD0
 
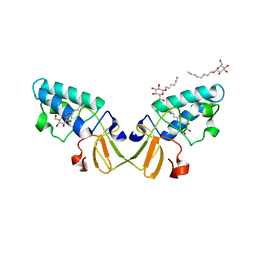 | |
9CAS
 
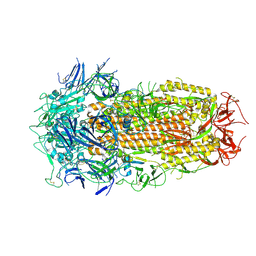 | |
6UTA
 
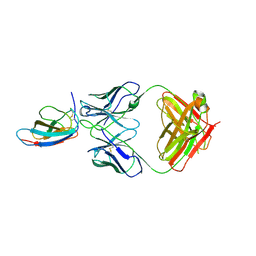 | | Crystal structure of Z004 iGL Fab in complex with ZIKV EDIII | | Descriptor: | Env, Z004 iGL Fab heavy chain, Z004 iGL Fab light chain | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UTE
 
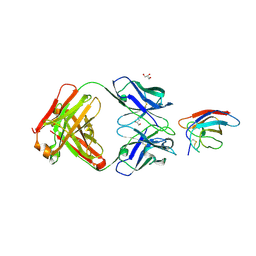 | | Crystal structure of Z032 Fab in complex with WNV EDIII | | Descriptor: | Envelope domain III, GLYCEROL, Z032 Fab heavy chain, ... | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8R7B
 
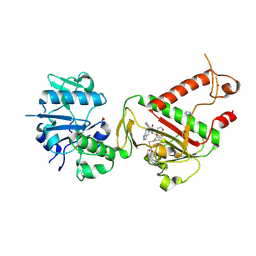 | | SARS-CoV-2 NSP14 in complex with SAH and TDI-015051 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Meyer, C, Garzia, A, Miller, M, Huggins, D.J, Myers, R.W, Liverton, N, Kargman, S, Nitsche, J, Ganichkin, O, Steinbacher, S, Meinke, P.T, Tuschl, T. | | Deposit date: | 2023-11-24 | | Release date: | 2024-10-23 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Small-molecule inhibition of SARS-CoV-2 NSP14 RNA cap methyltransferase.
Nature, 637, 2025
|
|
3P5O
 
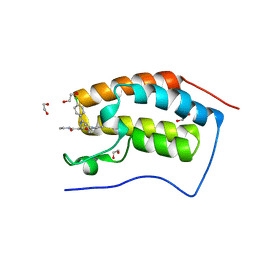 | | Crystal Structure of the First Bromodomain of Human Brd4 in complex with IBET inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2010-10-09 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Suppression of inflammation by a synthetic histone mimic
Nature, 468, 2010
|
|
7UAP
 
 | |
7UAR
 
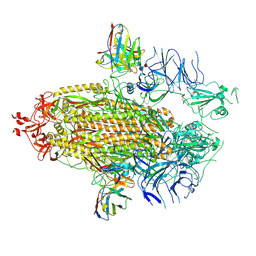 | |
7UAQ
 
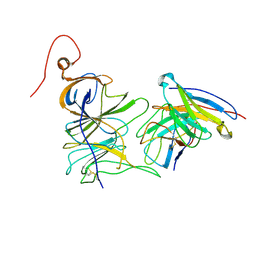 | | Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1520 Fab Heavy Chain, ... | | Authors: | Barnes, C.O. | | Deposit date: | 2022-03-13 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins.
Immunity, 55, 2022
|
|
6VJT
 
 | |
7LSE
 
 | |
7LSF
 
 | |
7LSG
 
 | |
1BT7
 
 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
5T5W
 
 | | Structure of an affinity matured lambda-IFN/IFN-lambdaR1/IL-10Rbeta receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon lambda receptor 1, Interferon lambda-3, ... | | Authors: | Mendoza, J.L, Jude, K.M, Garcia, K.C. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | The IFN-lambda-IFN-lambda R1-IL-10R beta Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity.
Immunity, 46, 2017
|
|
5VIC
 
 | | Crystal structure of anti-Zika antibody Z004 bound to DENV-1 Envelope protein DIII | | Descriptor: | Dengue 1 Envelope DIII domain, Fab heavy chain, Fab light chain | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
