9BBU
 
 | | SARS-CoV-2 Mpro in complex with compound 6h inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, N~2~-[4-(5-chloropyridin-3-yl)benzoyl]-N-{(1Z,2S)-1-imino-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Nayak, A, Afsar, M, Nayak, D, Olsen, S.K. | | Deposit date: | 2024-04-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 Mpro in complex with compound 6h inhibitor
To Be Published
|
|
9BBV
 
 | |
6C3R
 
 | |
6QEC
 
 | | DNA binding domain of LUX ARRYTHMO in complex with DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*GP*AP*AP*TP*AP*T*TP*AP*TP*AP*TP*TP*CP*GP*AP*A)-3'), GLYCEROL, Transcription factor LUX | | Authors: | Zubieta, C, Nayak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanisms of Evening Complex activity inArabidopsis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4O
 
 | | Solution structure of the Orange domain from human protein HES1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Fan, J.S, Nayak, A, Swaminathan, K. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induction of Transcriptional Inhibitor Hairy and Enhancer of Split Homolog-1 and the Related Repression of Tumor-Suppressor Thioredoxin-Interacting Protein Are Important Components of Cell-Transformation Program Imposed by Oncogenic Kinase Nucleophosmin-Anaplastic Lymphoma Kinase.
Am J Pathol, 2022
|
|
9BBT
 
 | | SARS-CoV-2 Mpro in complex with compound 6f inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2R)-1-imino-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-3',4'-dimethoxy[1,1'-biphenyl]-4-carboxamide | | Authors: | Afsar, M, Bury, P, Olsen, S.K, Nayak, A. | | Deposit date: | 2024-04-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | SARS-CoV-2 Mpro in complex with compound 6f inhibitor
To Be Published
|
|
9BBW
 
 | |
9BBS
 
 | | SARS-CoV-2 Mpro in complex with compound 6d inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2R)-1-imino-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4'-methoxy[1,1'-biphenyl]-4-carboxamide | | Authors: | Afsar, M, Bury, P, Olsen, S.K, Nayak, A. | | Deposit date: | 2024-04-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 Mpro in complex with compound 6d inhibitor
To Be Published
|
|
9BBX
 
 | |
8SEB
 
 | | Cryo-EM structure of a single loaded human UBA7-UBE2L6-ISG15 adenylate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SE9
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SEA
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 1) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8SV8
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex from a composite map | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
8FY1
 
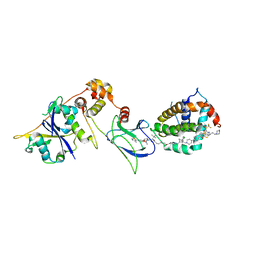 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY0
 
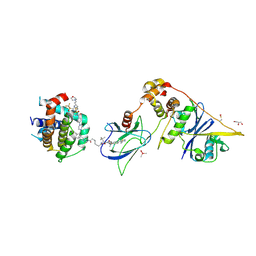 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | Descriptor: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | Authors: | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY2
 
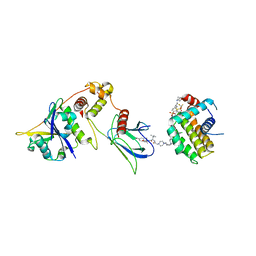 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
7SOL
 
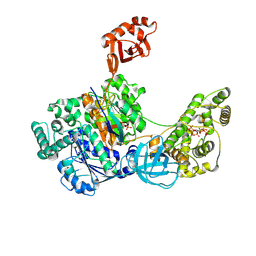 | | Crystal Structures of the bispecific ubiquitin/FAT10 activating enzyme, Uba6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Ubiquitin, ... | | Authors: | Olsen, S.K, Gao, F, Lv, Z. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25000644 Å) | | Cite: | Crystal structures reveal catalytic and regulatory mechanisms of the dual-specificity ubiquitin/FAT10 E1 enzyme Uba6.
Nat Commun, 13, 2022
|
|
7JRQ
 
 | | Crystallographically Characterized De Novo Designed Mn-Diphenylporphyrin Binding Protein | | Descriptor: | CHLORIDE ION, GLYCEROL, MPP1, ... | | Authors: | Mann, S.I, DeGrado, W.F. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De Novo Design, Solution Characterization, and Crystallographic Structure of an Abiological Mn-Porphyrin-Binding Protein Capable of Stabilizing a Mn(V) Species.
J.Am.Chem.Soc., 143, 2021
|
|
5LXU
 
 | | Structure of the DNA-binding domain of LUX ARRHYTHMO | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*TP*AP*TP*CP*TP*TP*AP*GP*AP*TP*AP*CP*GP*CP*A)-3'), Transcription factor LUX | | Authors: | Zubieta, C, Nanao, M.H. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of the DNA-binding domain of LUX ARRHYTHMO
To Be Published
|
|
7SND
 
 | | Pacifastin related protease inhibitors | | Descriptor: | GLYCEROL, PHOSPHATE ION, Pacifastin-related peptide | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
7SNC
 
 | | Pacifastin related protease inhibitors | | Descriptor: | Protease inhibitor | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-10-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
5MJA
 
 | | Kinase domain of human EphB1 bound to a quinazoline-based inhibitor | | Descriptor: | 2-chloranyl-~{N}-[4-[(2-chloranyl-5-oxidanyl-phenyl)amino]quinazolin-7-yl]ethanamide, Ephrin type-B receptor 1, SULFATE ION | | Authors: | Kung, A, Schimpl, M, Chen, Y.-C, Overman, R.C, Zhang, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A Chemical-Genetic Approach to Generate Selective Covalent Inhibitors of Protein Kinases.
ACS Chem. Biol., 12, 2017
|
|
5MJB
 
 | | Kinase domain of human EphB1, G703C mutant, covalently bound to a quinazoline-based inhibitor | | Descriptor: | 2-chloranyl-~{N}-[4-[(2-chloranyl-5-oxidanyl-phenyl)amino]quinazolin-7-yl]ethanamide, Ephrin type-B receptor 1, SULFATE ION | | Authors: | Kung, A, Schimpl, M, Chen, Y.-C, Overman, R.C, Zhang, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical-Genetic Approach to Generate Selective Covalent Inhibitors of Protein Kinases.
ACS Chem. Biol., 12, 2017
|
|
7SAP
 
 | | The CTI-homolog pacifastin | | Descriptor: | GLYCEROL, Serine protease inhibitor I/II-like Protein | | Authors: | Gewe, M.M, Strong, R.K. | | Deposit date: | 2021-09-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ex silico engineering of cystine-dense peptides yielding a potent bispecific T cell engager.
Sci Transl Med, 14, 2022
|
|
8TB1
 
 | |
