5DQL
 
 | |
6WSI
 
 | | Intact cis-2,3-epoxysuccinic acid bound to Isocitrate Lyase-1 from Mycobacterium tuberculosis | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Covalent Inactivation of Mycobacterium tuberculosis Isocitrate Lyase by cis -2,3-Epoxy-Succinic Acid.
Acs Chem.Biol., 16, 2021
|
|
6UX6
 
 | | Cruzain covalently bound by a vinylsulfone compound | | Descriptor: | Cruzipain, GLYCEROL, Nalpha-[(benzyloxy)carbonyl]-N-[(2S)-1-phenyl-4-(phenylsulfonyl)butan-2-yl]-L-phenylalaninamide | | Authors: | Zhai, X, Tang, S, Chenna, B.C, Meek, T.D, Sacchettini, J.C. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Cruzain covalently bound by a vinylsulfone compound
To Be Published
|
|
6VB9
 
 | | Covalent adduct of cis-2,3-epoxysuccinic acid with Isocitrate Lyase-1 from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Isocitrate lyase, ... | | Authors: | Krieger, I.V, Pham, T.V, Meek, T.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Covalent Inactivation of Mycobacterium tuberculosis Isocitrate Lyase by cis -2,3-Epoxy-Succinic Acid.
Acs Chem.Biol., 16, 2021
|
|
2ADU
 
 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | Descriptor: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
2OAZ
 
 | | Human Methionine Aminopeptidase-2 Complexed with SB-587094 | | Descriptor: | COBALT (II) ION, N-(2-ISOPROPYLPHENYL)-3-[(2-THIENYLMETHYL)THIO]-1H-1,2,4-TRIAZOL-5-AMINE, human Methionine Amino Peptidase 2 | | Authors: | Marino Jr, J.P, Fisher, P.W, Hofmann, G.A, Kirkpatrick, R, Janson, C.A, Johnson, R.K, Ma, C, Mattern, M, Meek, T.D, Ryan, D, Schulz, C, Smith, W.W, Tew, D.G, Tomazek Jr, T.A, Veber, D.F, Xiong, W.C, Yamamoto, Y, Yamashita, K, Yang, G, Thompson, S.K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of methionine aminopeptidase-2 based on a 1,2,4-triazole pharmacophore.
J.Med.Chem., 50, 2007
|
|
8FWY
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to the dead-end complex xanthine and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, XANTHINE | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX3
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-GP, showing the structure of the complete active site in its open conformation | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX1
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (R)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX0
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (S)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX2
 
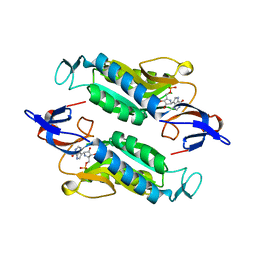 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-HP | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FWZ
 
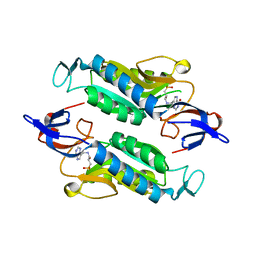 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Hydroxypropyl-Lin-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, [(2R)-2-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}propyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
7RB1
 
 | | Isocitrate Lyase-1 from Mycobacterium tuberculosis covalently modified by 5-descarboxy-5-nitro-D-isocitric acid | | Descriptor: | (3E)-3-(hydroxyimino)propanoic acid, GLYCEROL, GLYOXYLIC ACID, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T, Sacchettini, J.C. | | Deposit date: | 2021-07-05 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Based Inactivation of Mycobacterium tuberculosis Isocitrate Lyase 1 by (2 R ,3 S )-2-Hydroxy-3-(nitromethyl)succinic acid.
J.Am.Chem.Soc., 143, 2021
|
|
1HPS
 
 | | RATIONAL DESIGN, SYNTHESIS AND CRYSTALLOGRAPHIC ANALYSIS OF A HYDROXYETHYLENE-BASED HIV-1 PROTEASE INHIBITOR CONTAINING A HETEROCYCLIC P1'-P2' AMIDE BOND ISOSTERE | | Descriptor: | 2-[(1R,3S,4S)-1-BENZYL-4-[N-(BENZYLOXYCARBONYL)-L-VALYL]AMINO-3-PHENYLPENTYL]-4(5)-(2-METHYLPROPIONYL)IMIDAZOLE, HIV-1 PROTEASE | | Authors: | Abdel-Meguid, S, Zhao, B. | | Deposit date: | 1994-05-24 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis, and crystallographic analysis of a hydroxyethylene-based HIV-1 protease inhibitor containing a heterocyclic P1'--P2' amide bond isostere.
J.Med.Chem., 37, 1994
|
|
1SBG
 
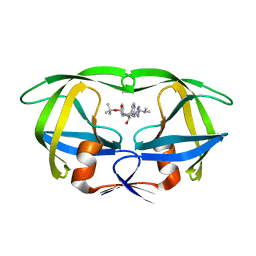 | |
1AAQ
 
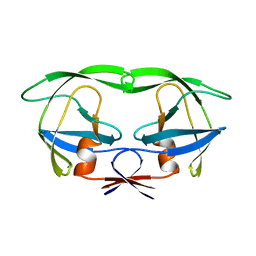 | |
1HOS
 
 | |
4OEM
 
 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
4OEL
 
 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Dipeptidyl peptidase 1 Heavy chain, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
3STR
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine hydroxamic acid | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-[2-(hydroxyamino)-2-oxoethyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
3SW8
 
 | |
3SVJ
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine amide | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-methyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-12 | | Release date: | 2011-07-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
7M2P
 
 | |
