7KPB
 
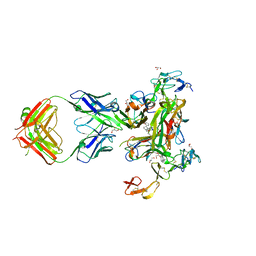 | | Human TNF-alpha TNFR1 complex bound to conformationally selective antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(1-{[2-(difluoromethoxy)phenyl]methyl}-2-{[3-(2-oxopyrrolidin-1-yl)phenoxy]methyl}-1H-benzimidazol-6-yl)pyridin-2(1H)-one, Fab1974 - Heavy Chain, ... | | Authors: | Fox III, D, Conrady, D.G, Lowe, M, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A conformation-selective monoclonal antibody against a small molecule-stabilised signalling-deficient form of TNF
Nat Commun, 12, 2021
|
|
1BU8
 
 | | RAT PANCREATIC LIPASE RELATED PROTEIN 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PANCREATIC LIPASE RELATED PROTEIN 2) | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-09-14 | | Release date: | 1998-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of rat pancreatic lipase-related protein 2.
J.Biol.Chem., 273, 1998
|
|
4TVJ
 
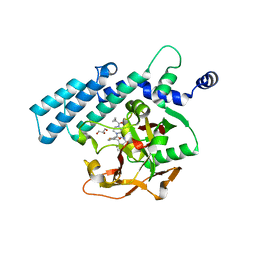 | | HUMAN ARTD2 (PARP2) - CATALYTIC DOMAIN IN COMPLEX WITH OLAPARIB | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Pinto, A.F, Schuler, H. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4UXB
 
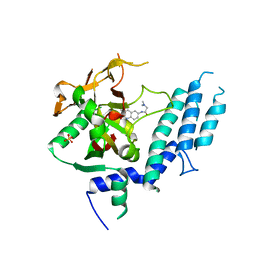 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, POLY ADP-RIBOSE POLYMERASE 1, SULFATE ION | | Authors: | Tresaugues, L, Thorsell, A.G, Karlberg, T, Schuler, H. | | Deposit date: | 2014-08-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
8W01
 
 | |
8W04
 
 | |
4UND
 
 | | HUMAN ARTD1 (PARP1) - CATALYTIC DOMAIN IN COMPLEX WITH INHIBITOR TALAZOPARIB | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, POLY [ADP-RIBOSE] POLYMERASE 1, SODIUM ION | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Klepsch, M, Pinto, A.F, Tresaugues, L, Moche, M, Schuler, H. | | Deposit date: | 2014-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
8U48
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 D161A-E163A inactive Endoglycosidase in complex with high-mannose N-glycan (Man9GlcNAc2) substrate | | Descriptor: | Endo-beta-N-acetylglucosaminidase, PHOSPHATE ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sastre, D.E, Sultana, N, Navarro, M.V.A.S, Sundberg, E.J. | | Deposit date: | 2023-09-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
8U46
 
 | |
8U47
 
 | |
8U9F
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 in complex with NaI | | Descriptor: | 1,2-ETHANEDIOL, Endo-beta-N-acetylglucosaminidase, IODIDE ION, ... | | Authors: | Sastre, D.E, Navarro, M.V.A.S, Sundberg, E.J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
5LX6
 
 | |
8VFU
 
 | |
8UEN
 
 | |
8URA
 
 | |
4R5W
 
 | | Human artd1 (parp1) - catalytic domain in complex with inhibitor xav939 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J.Med.Chem., 60, 2017
|
|
4R6E
 
 | | Human artd1 (parp1) - catalytic domain in complex with inhibitor niraparib | | Descriptor: | 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Karlberg, T, Thorsell, A.G, Brock, J, Schuler, H. | | Deposit date: | 2014-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J.Med.Chem., 60, 2017
|
|
4RV6
 
 | |
