4Z0B
 
 | | Crystal Structure of the Fab Fragment of Anti-ofloxacin Antibody and Exploration Its Receptor Binding Site | | Descriptor: | PHOSPHATE ION, antibody heavy chain, antibody light chain | | Authors: | He, K, Du, X, Sheng, W, Zhou, X, Wang, J, Wang, S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-ofloxacin Antibody and Exploration of Its Specific Binding.
J.Agric.Food Chem., 64, 2016
|
|
2P16
 
 | | Factor Xa in Complex with the Inhibitor APIXABAN (BMS-562247) AKA 1-(4-METHOXYPHENYL)-7-OXO-6-(4-(2-OXO-1-PIPERIDINYL)PHENYL)-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3, 4-C]PYRIDINE-3-CARBOXAMIDE | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, CALCIUM ION, Coagulation factor X (EC 3.4.21.6) (Stuart factor) (Stuart-Prower factor) | | Authors: | Alexander, R. | | Deposit date: | 2007-03-02 | | Release date: | 2007-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 1-(4-Methoxyphenyl)-7-oxo-6-(4-(2-oxopiperidin-1-yl)phenyl)-4,5,6,7-tetrahydro- 1H-pyrazolo[3,4-c]pyridine-3-carboxamide (Apixaban, BMS-562247), a Highly Potent, Selective, Efficacious, and Orally Bioavailable Inhibitor of Blood Coagulation Factor Xa.
J.Med.Chem., 50, 2007
|
|
2FZZ
 
 | | Factor Xa in complex with the inhibitor 1-(3-amino-1,2-benzisoxazol-5-yl)-6-(2'-(((3r)-3-hydroxy-1-pyrrolidinyl)methyl)-4-biphenylyl)-3-(trifluoromethyl)-1,4,5,6-tetrahydro-7h-pyrazolo[3,4-c]pyridin-7-one | | Descriptor: | 1-(3-AMINO-1,2-BENZISOXAZOL-5-YL)-6-(2'-{[(3R)-3-HYDROXYPYRROLIDIN-1-YL]METHYL}BIPHENYL-4-YL)-3-(TRIFLUOROMETHYL)-1,4,5,6-TETRAHYDRO-7H-PYRAZOLO[3,4-C]PYRIDIN-7-ONE, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2006-02-10 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1-[3-Aminobenzisoxazol-5'-yl]-3-trifluoromethyl-6-[2'-(3-(R)-hydroxy-N-pyrrolidinyl)methyl-[1,1']-biphen-4-yl]-1,4,5,6-tetrahydropyrazolo-[3,4-c]-pyridin-7-one (BMS-740808) a highly potent, selective, efficacious, and orally bioavailable inhibitor of blood coagulation factor Xa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3CEN
 
 | |
3CS7
 
 | | FACTOR XA IN COMPLEX WITH THE INHIBITOR 1-(4-methoxyphenyl)-6-(4-(1-(pyrrolidin-1-ylmethyl)cyclopropyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one | | Descriptor: | 1-(4-methoxyphenyl)-6-(4-(1-(pyrrolidin-1-ylmethyl)cyclopropyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one, CALCIUM ION, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Achieving structural diversity using the perpendicular conformation of alpha-substituted phenylcyclopropanes to mimic the bioactive conformation of ortho-substituted biphenyl P4 moieties: discovery of novel, highly potent inhibitors of Factor Xa.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2G00
 
 | | Factor Xa in complex with the inhibitor 3-(6-(2'-((dimethylamino)methyl)-4-biphenylyl)-7-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-c]pyridin-1-yl)benzamide | | Descriptor: | 3-[6-{2'-[(DIMETHYLAMINO)METHYL]BIPHENYL-4-YL}-7-OXO-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN-1-YL]BENZAMIDE, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2006-02-10 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of potent, efficacious, and orally bioavailable inhibitors of blood coagulation factor Xa with neutral P1 moieties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5HNQ
 
 | |
5HN2
 
 | |
5HNJ
 
 | |
5YKO
 
 | |
5YKN
 
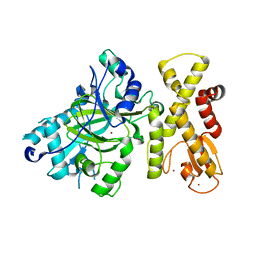 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | Descriptor: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | Authors: | Yang, Z, Du, J. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
6JNL
 
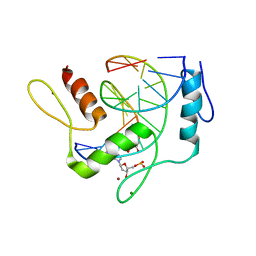 | | REF6 ZnF2-4-NAC004 complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
6JNM
 
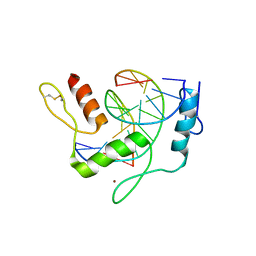 | | REF6 ZnF2-4-NAC004-mC3 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*(5CM)P*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
6JNN
 
 | | REF6 ZnF2-4-NAC004-mC1 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*(5CM)P*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
8HGO
 
 | | The EGF-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGP
 
 | | The EREG-bound EGFR/HER2 ectodomain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HGS
 
 | | The EGF-bound EGFR ectodomain homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Pro-epidermal growth factor, ... | | Authors: | Zhang, Z, Bai, X. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure and dynamics of the EGFR/HER2 heterodimer.
Cell Discov, 9, 2023
|
|
8HR1
 
 | | Cryo-EM structure of SSX1 bound to the unmodified nucleosome at a resolution of 3.02 angstrom | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Zebin, T, Ai, H.S, Ziyu, X, Man, P, Liu, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQY
 
 | | Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom | | Descriptor: | DNA (136-MER), DNA (137-MER), Histone H2A type 1-B/E, ... | | Authors: | Zebin, T, Ai, H.S, Ziyu, X, GuoChao, C, Man, P, Liu, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7DNN
 
 | | Crystal structure of the AgCarB2-C2 complex with homoorientin | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-6-[(2S,3R,4R,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-5,7-bis(oxidanyl)chromen-4-one, AP_endonuc_2 domain-containing protein, AgCarC2, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DNM
 
 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
