7YWV
 
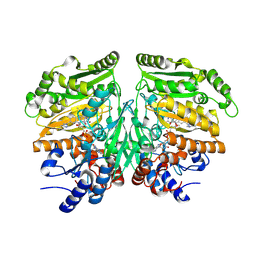 | | Eugenol oxidase from rhodococcus jostii: mutant S81H, D151E, A423M, H434Y, S394V, Q425S, I445D, S518P | | 分子名称: | 2-methoxy-4-(prop-2-en-1-yl)phenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Alvigini, L, Mattevi, A. | | 登録日 | 2022-02-14 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure- and computational-aided engineering of an oxidase to produce isoeugenol from a lignin-derived compound.
Nat Commun, 13, 2022
|
|
5JIE
 
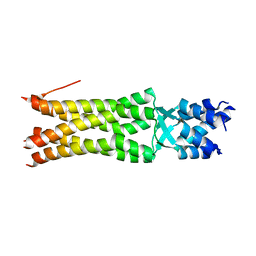 | |
3GIZ
 
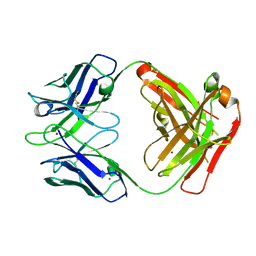 | | Crystal structure of the Fab fragment of anti-CD20 antibody Ofatumumab | | 分子名称: | Fab fragment of anti-CD20 antibody Ofatumumab, heavy chain, light chain, ... | | 著者 | Du, J, Yang, H, Ding, J. | | 登録日 | 2009-03-07 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the Fab fragment of therapeutic antibody Ofatumumab provides insights into the recognition mechanism with CD20
Mol.Immunol., 46, 2009
|
|
7TTM
 
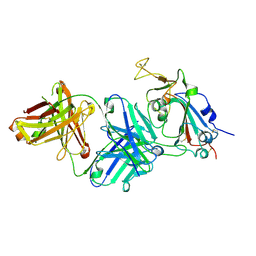 | |
7TTX
 
 | |
7TTY
 
 | |
5CX3
 
 | | Crystal structure of FYCO1 LIR in complex with LC3A | | 分子名称: | FYVE and coiled-coil domain-containing protein 1, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Cheng, X, Pan, L. | | 登録日 | 2015-07-28 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Autophagy, 12, 2016
|
|
6NF2
 
 | |
7N5H
 
 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | 分子名称: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Casner, R.G, Cerutti, G, Shapiro, L. | | 登録日 | 2021-06-05 | | 公開日 | 2021-11-03 | | 最終更新日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
2PVX
 
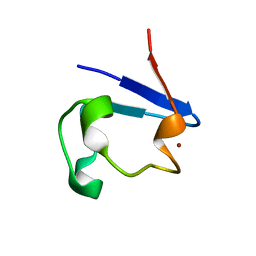 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | 分子名称: | Rubredoxin, ZINC ION | | 著者 | Wang, L, LeMaster, D.M, Hernandez, G, Li, H. | | 登録日 | 2007-05-10 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin
Bmc Struct.Biol., 7, 2007
|
|
4WD8
 
 | |
8G59
 
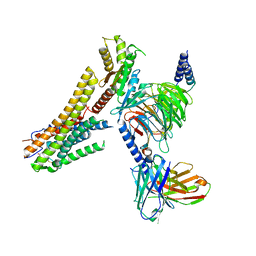 | | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | | 分子名称: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | 登録日 | 2023-02-12 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
3STW
 
 | |
3STX
 
 | |
3STY
 
 | |
7MHS
 
 | | Structure of p97 (subunits A to E) with substrate engaged | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Xu, Y, Han, H, Cooney, I, Hill, C.P, Shen, P.S. | | 登録日 | 2021-04-15 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Active conformation of the p97-p47 unfoldase complex.
Nat Commun, 13, 2022
|
|
6NZ7
 
 | |
6OT1
 
 | |
6OSY
 
 | |
6PZV
 
 | |
7LS9
 
 | |
7LSS
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-7 variable heavy chain, ... | | 著者 | Rapp, M, Shapiro, L. | | 登録日 | 2021-02-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies.
Structure, 29, 2021
|
|
7L5B
 
 | |
3PMT
 
 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | 分子名称: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | 著者 | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-18 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-05-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
5KF4
 
 | |
