1FI5
 
 | | NMR STRUCTURE OF THE C TERMINAL DOMAIN OF CARDIAC TROPONIN C BOUND TO THE N TERMINAL DOMAIN OF CARDIAC TROPONIN I. | | Descriptor: | CALCIUM ION, PROTEIN (TROPONIN C) | | Authors: | Gasmi-Seabrook, G.M, Howarth, J.W, Finley, N, Abusamhadneh, E, Gaponenko, V, Brito, R.M, Solaro, R.J, Rosevear, P.R. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the C-terminal domain of cardiac troponin C free and bound to the N-terminal domain of cardiac troponin I.
Biochemistry, 38, 1999
|
|
7RSE
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSC
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
2LQI
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2L04
 
 | | The Solution Structure of the C-terminal Ig-like Domain of the Bacteriophage Lambda Tail Tube Protein | | Descriptor: | Major tail protein V | | Authors: | Pell, L.G, Gasmi-Seabrook, G.M.C, Donaldson, L.W, Howell, P, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the C-Terminal Ig-like Domain of the Bacteriophage l Tail Tube Protein.
J.Mol.Biol., 403, 2010
|
|
2LQH
 
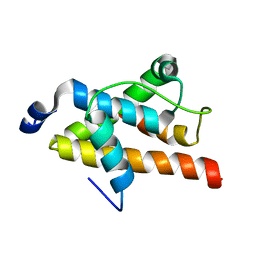 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6W4E
 
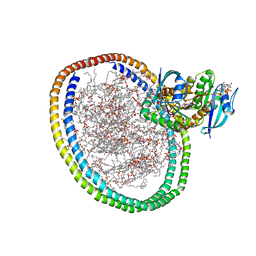 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4KEJ
 
 | |
4KEI
 
 | |
4KEK
 
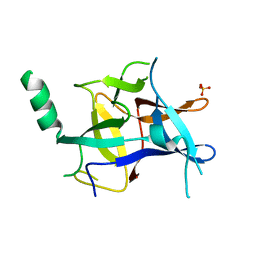 | |
2MC2
 
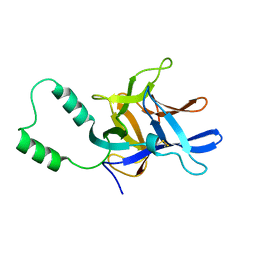 | |
2MSD
 
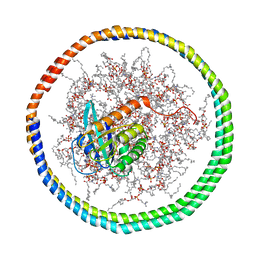 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
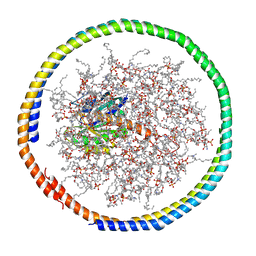 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MAJ
 
 | |
2MAK
 
 | |
1SCV
 
 | |
7KMR
 
 | |
7SCW
 
 | | KRAS full length wild-type in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
7SCX
 
 | | KRAS full-length G12V in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
7JII
 
 | | HRAS A59E GDP | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIF
 
 | | HRAS A59T GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIH
 
 | | HRAS A59E GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIG
 
 | | HRAS A59T GppNHp crystal 2 | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
