1ZVI
 
 | | Rat Neuronal Nitric Oxide Synthase Oxygenase Domain | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric-oxide synthase, brain, ... | | Authors: | Matter, H, Kumar, H.S, Fedorov, R, Frey, A, Kotsonis, P, Hartmann, E, Frohlich, L.G, Reif, A, Pfleiderer, W, Scheurer, P, Ghosh, D.K, Schlichting, I, Schmidt, H.H. | | Deposit date: | 2005-06-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Isoform-Specific Inhibitors Targeting the Tetrahydrobiopterin Binding Site of Human Nitric Oxide Synthases.
J.Med.Chem., 48, 2005
|
|
4EL6
 
 | | Crystal structure of IPSE/alpha-1 from Schistosoma mansoni eggs | | Descriptor: | IL-4-inducing protein | | Authors: | Mayerhofer, H, Meyer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Sattler, M, Schramm, G, Mueller-Dieckmann, J. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A Crystallin Fold in the Interleukin-4-inducing Principle of Schistosoma mansoni Eggs (IPSE/ alpha-1) Mediates IgE Binding for Antigen-independent Basophil Activation.
J.Biol.Chem., 290, 2015
|
|
4BA9
 
 | | The structural basis for the coordination of Y-family Translesion DNA Polymerases by Rev1 | | Descriptor: | DNA POLYMERASE KAPPA, DNA REPAIR PROTEIN REV1, MAGNESIUM ION, ... | | Authors: | Grummitt, C.G, Kilkenny, M.L, Frey, A, Roe, S.M, Oliver, A.W, Sale, J.E, Pearl, L.H. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Structural Basis for the Coordination of Y- Family Translesion DNA Polymerases by Rev1
To be Published
|
|
4AKA
 
 | | IPSE alpha-1, an IgE-binding crystallin | | Descriptor: | IL-4-INDUCING PROTEIN | | Authors: | Meyer, N.H, Mayerhofer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Mueller-Dieckmann, J, Sattler, M, Scharmm, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Crystallin Fold in the Interleukin-4-Inducing Principle of Schistosoma Mansoni Eggs (Ipse/Alpha-1) Mediates Ige Binding for Antigen-Independent Basophil Activation
J.Biol.Chem., 290, 2015
|
|
1ZVL
 
 | | Rat Neuronal Nitric Oxide Synthase Oxygenase Domain complexed with natural substrate L-Arg. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, Nitric-oxide synthase, ... | | Authors: | Matter, H, Kumar, H.S, Fedorov, R, Frey, A, Kotsonis, P, Hartmann, E, Frohlich, L.G, Reif, A, Pfleiderer, W, Scheurer, P, Ghosh, D.K, Schlichting, I, Schmidt, H.H. | | Deposit date: | 2005-06-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Isoform-Specific Inhibitors Targeting the Tetrahydrobiopterin Binding Site of Human Nitric Oxide Synthases.
J.Med.Chem., 48, 2005
|
|
2QCE
 
 | | Crystal structure of the orotidine-5'-monophosphate decarboxylase domain of human UMP synthase bound to sulfate, glycerol, and chloride | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Wittmann, J, Rudolph, M. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of the human orotidine-5'-monophosphate decarboxylase support a covalent mechanism and provide a framework for drug design.
Structure, 16, 2008
|
|
2QCC
 
 | |
2QCL
 
 | |
2QCM
 
 | |
2QCH
 
 | |
2QCG
 
 | |
2QCN
 
 | |
2QCF
 
 | |
2QCD
 
 | |
1DMJ
 
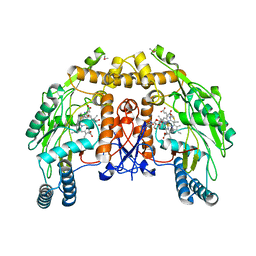 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 5,6-CYCLIC-TETRAHYDROPTERIDINE | | Descriptor: | 7-AMINO-3,3A,4,5-TETRAHYDRO-8H-2-OXA-5,6,8,9B-TETRAAZA-CYCLOPENTA[A]NAPHTHALENE-1,9-DIONE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
7M8H
 
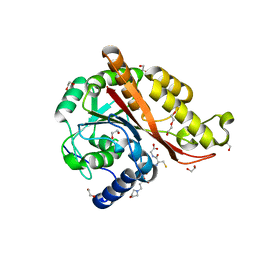 | | Structure of Memo1 C244S metal binding site mutant at 1.75A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2021-03-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
6M3C
 
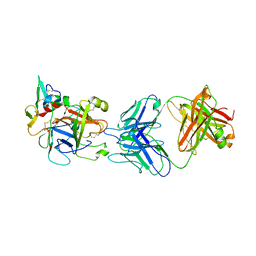 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
6M3B
 
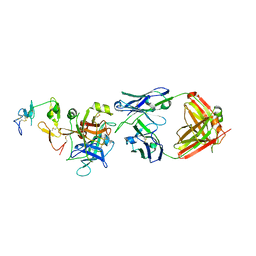 | | hAPC-c25k23 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, ... | | Authors: | Wang, X, Li, L, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
1LMB
 
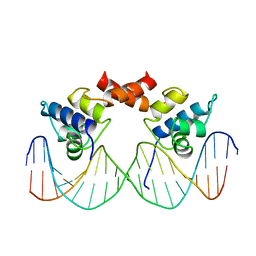 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
1LRP
 
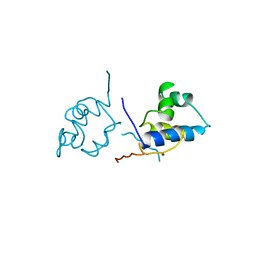 | |
7KQ8
 
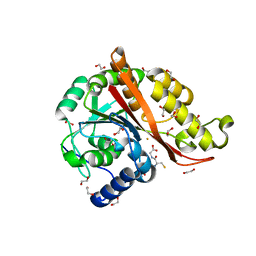 | | Structure of iron bound MEMO1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2020-11-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
7L5C
 
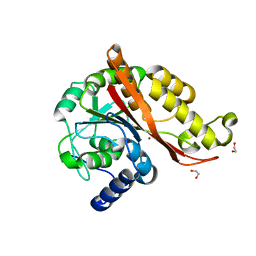 | | Structure of copper bound MEMO1 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, GLYCEROL, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
8R6X
 
 | |
7K7J
 
 | | EphB6 receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 6 | | Authors: | Goldgur, Y, Himanen, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of the EphB6 receptor ectodomain.
Plos One, 16, 2021
|
|
