2RER
 
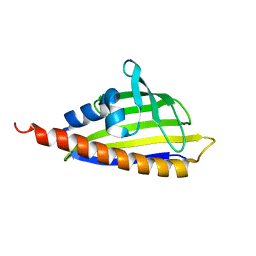 | |
2RES
 
 | | Tetracenomycin ARO/CYC mutant R69A | | Descriptor: | Multifunctional cyclase-dehydratase-3-O-methyl transferase tcmN | | Authors: | Ames, B.D, Tsai, S.C. | | Deposit date: | 2007-09-26 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4KEH
 
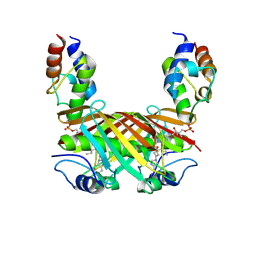 | | Crosslinked Crystal Structure of Type II Fatty Synthase Dehydratase, FabA, and Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, N-{3-[DIHYDROXY(NONYL)-LAMBDA~4~-SULFANYL]PROPYL}-N~3~-[(2R)-2-HYDROXY-3,3-DIMETHYL-4-(PHOSPHONOOXY)BUTANOYL]-BETA-ALANINAMIDE, N-{3-[dihydroxy(nonyl)-lambda~4~-sulfanyl]propyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Nguyen, C, Haushalter, R, Finzel, K, Leong, J, Le, B.C, Burkart, M, Tsai, S.C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-12-25 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Trapping the dynamic acyl carrier protein in fatty acid biosynthesis.
Nature, 505, 2014
|
|
1KEZ
 
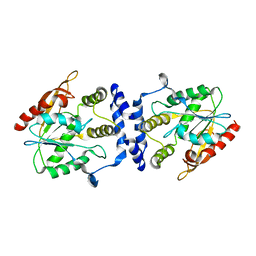 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4DC0
 
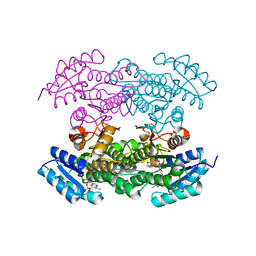 | |
4DBZ
 
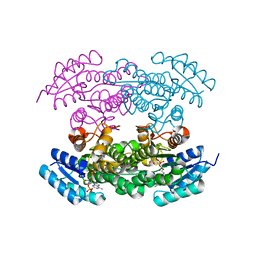 | |
4DC1
 
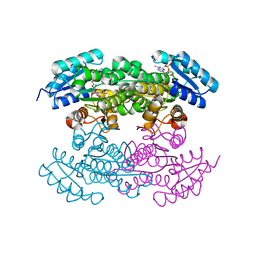 | |
3B6Z
 
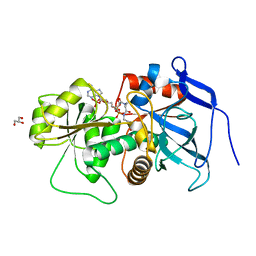 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B70
 
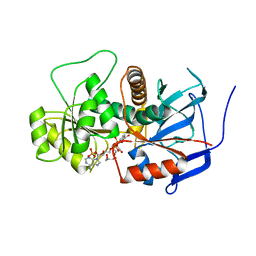 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RI3
 
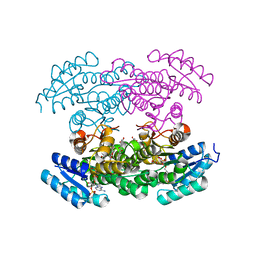 | |
3SJU
 
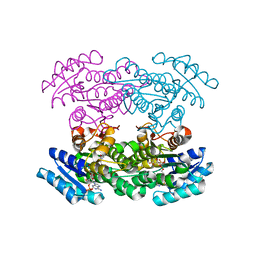 | | Hedamycin Polyketide Ketoreductase bound to NADPH | | Descriptor: | Keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Javidpour, P, Tsai, S.-C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Studies of the Hedamycin Type II Polyketide Ketoreductase (HedKR): Molecular Basis of Stereo- and Regiospecificities.
Biochemistry, 50, 2011
|
|
3TVR
 
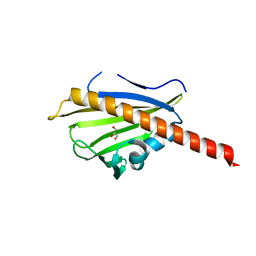 | |
3TVQ
 
 | | Crystal structure of TCM Aro/Cyc complexed with trans-dihidroquercetin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, Multifunctional cyclase-dehydratase-3-O-methyl transferase tcmN | | Authors: | Ames, B.D, Lee, M.-Y, Tsai, S.-C. | | Deposit date: | 2011-09-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
3TL1
 
 | | Crystal structure of the Streptomyces coelicolor WhiE ORFVI polyketide aromatase/cyclase | | Descriptor: | 6,7,9-trihydroxy-3-methyl-1H-benzo[g]isochromen-1-one, GLYCEROL, Polyketide cyclase | | Authors: | Lee, M.-Y, Ames, B.D, Zhang, W, Tang, Y, Tsai, S.-C. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
3HF1
 
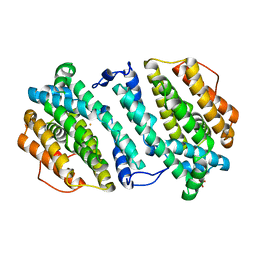 | | Crystal structure of human p53R2 | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase subunit M2 B, SULFATE ION | | Authors: | Smith, P, Zhou, B, Yuan, Y.-C, Su, L, Tsai, S.-C, Yen, Y. | | Deposit date: | 2009-05-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A X-ray crystal structure of human p53R2, a p53-inducible ribonucleotide reductase .
Biochemistry, 48, 2009
|
|
3QRW
 
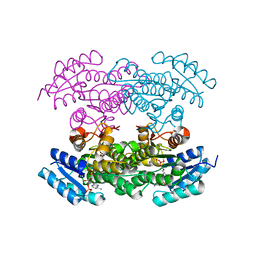 | |
1MZJ
 
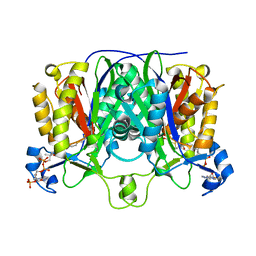 | | Crystal Structure of the Priming beta-Ketosynthase from the R1128 Polyketide Biosynthetic Pathway | | Descriptor: | ACETYL GROUP, Beta-ketoacylsynthase III, COENZYME A | | Authors: | Pan, H, Tsai, S.C, Meadows, E.S, Miercke, L.J.W, Keatinge-Clay, A, O'Connell, J, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-10-08 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the priming beta-ketosynthase from the
R1128 polyketide biosynthetic pathway
Structure, 10
|
|
1X7G
 
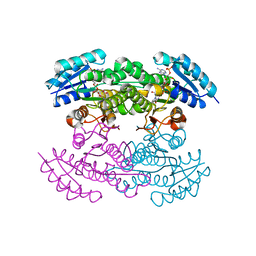 | | Actinorhodin Polyketide Ketoreductase, act KR, with NADP bound | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
1X7H
 
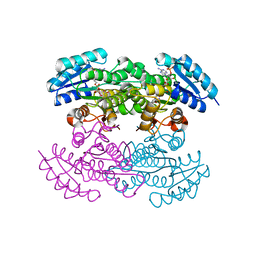 | | Actinorhodin Polyketide Ketoreductase, with NADPH bound | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
3IB9
 
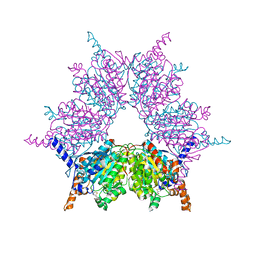 | | Propionyl-CoA Carboxylase Beta Subunit, D422L | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase complex B subunit, SULFATE ION | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3IAV
 
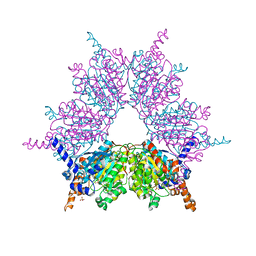 | | Propionyl-CoA Carboxylase Beta Subunit, D422V | | Descriptor: | Propionyl-CoA carboxylase complex B subunit, SULFATE ION | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3IBB
 
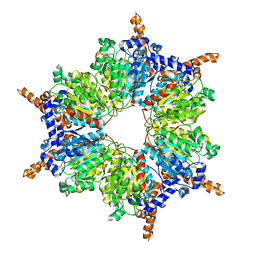 | | Propionyl-CoA Carboxylase Beta Subunit, D422A | | Descriptor: | Propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3ILS
 
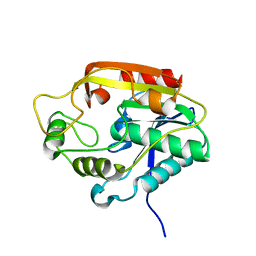 | | The Thioesterase Domain from PksA | | Descriptor: | Aflatoxin biosynthesis polyketide synthase | | Authors: | Korman, T.P. | | Deposit date: | 2009-08-07 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MFM
 
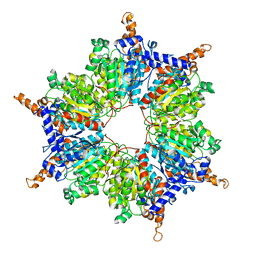 | | Crystal Structures and Mutational Analyses of Acyl-CoA Carboxylase Subunit of Streptomyces coelicolor | | Descriptor: | Propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Melgar, M.M. | | Deposit date: | 2010-04-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3NI3
 
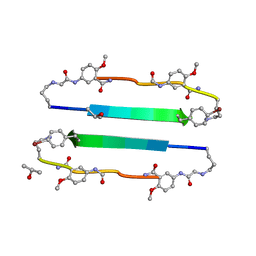 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
