8JNI
 
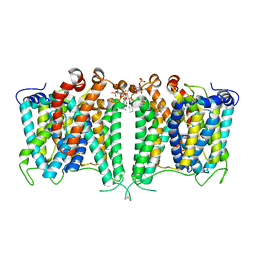 | | Structure of AE2 in complex with PIP2 | | Descriptor: | Anion exchange protein 2, CHLORIDE ION, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
3IN0
 
 | |
3IN2
 
 | |
3JT2
 
 | |
3JTB
 
 | |
7EVO
 
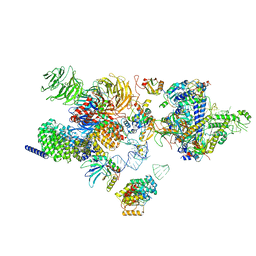 | | The cryo-EM structure of the human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
7EVN
 
 | | The cryo-EM structure of the DDX42-SF3b complex | | Descriptor: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
5TAB
 
 | |
5TBN
 
 | |
5U39
 
 | | Pseudomonas aeruginosa LpxC in complex with CHIR-090 | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
5U3B
 
 | | Pseudomonas aeruginosa LpxC in complex with NVS-LPXC-01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[(but-2-yn-1-yl)oxy]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
1NS1
 
 | |
2B29
 
 | | N-terminal domain of the RPA70 subunit of human replication protein A. | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Bochkareva, E, Kaustov, L, Ayed, A, Okorokov, A, Milner, J, Arrowsmith, C.H, Bochkarev, A. | | Deposit date: | 2005-09-18 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4L08
 
 | | Crystal structure of the maleamate amidase Ami(C149A) in complex with maleate from Pseudomonas putida S16 | | Descriptor: | Hydrolase, isochorismatase family, MALEIC ACID | | Authors: | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-31 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
7EPT
 
 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
4L07
 
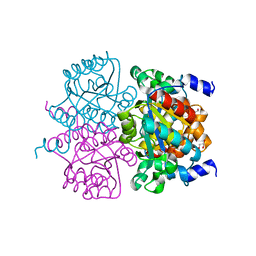 | | Crystal structure of the maleamate amidase Ami from Pseudomonas putida S16 | | Descriptor: | GLYCEROL, Hydrolase, isochorismatase family, ... | | Authors: | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
7KHL
 
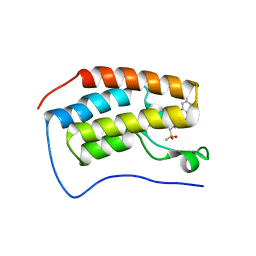 | | BRD4-BD1 Compound6 (methyl 4-(3,5-difluoropyridin-2-yl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxylate) | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, methyl 7-(3,5-difluoropyridin-2-yl)-2-methyl-10-[(methylsulfonyl)methyl]-3-oxo-3,4,6,7-tetrahydro-2H-2,4,7-triazadibenzo[cd,f]azulene-9-carboxylate | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.286 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
7KHH
 
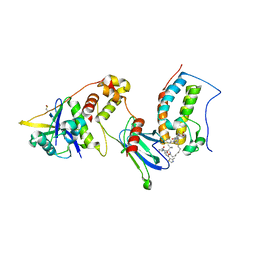 | | Ternary complex of VHL/BRD4-BD1/Compound9 (4-(3,5-difluoropyridin-2-yl)-N-(11-(((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-11-oxoundecyl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxamide) | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
8HMZ
 
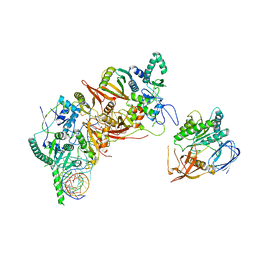 | | Cryo-EM structure of the human post-catalytic TSEN/pre-tRNA complex | | Descriptor: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | Authors: | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
8HMY
 
 | | Cryo-EM structure of the human pre-catalytic TSEN/pre-tRNA complex | | Descriptor: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | Authors: | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
6BD4
 
 | | Crystal structure of human apo-Frizzled4 receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Frizzled-4/Rubredoxin chimeric protein, OLEIC ACID, ... | | Authors: | Yang, S, Wu, Y, Pu, M, Chen, Y, Dong, S, Guo, Y, Han, G.Y, Stevens, R.C, Zhao, S, Xu, F. | | Deposit date: | 2017-10-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Frizzled 4 receptor in a ligand-free state.
Nature, 560, 2018
|
|
2F5J
 
 | | Crystal structure of MRG domain from human MRG15 | | Descriptor: | Mortality factor 4-like protein 1 | | Authors: | Zhang, P, Du, J, Ding, J. | | Deposit date: | 2005-11-26 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14
Protein Sci., 15, 2006
|
|
8BQU
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
2H06
 
 | |
2H07
 
 | |
