6XCF
 
 | |
2PQS
 
 | | Crystal Structure of the Bovine Lactadherin C2 Domain | | Descriptor: | Lactadherin | | Authors: | Huang, M, Furie, B.C. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the bovine lactadherin C2 domain, a membrane binding motif, shows similarity to the C2 domains of factor V and factor VIII.
J.Mol.Biol., 371, 2007
|
|
3LAQ
 
 | | Structure-based engineering of species selectivity in the uPA-uPAR interaction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator | | Authors: | Huang, M. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based engineering of species selectivity in the interaction between urokinase and its receptor: implication for preclinical cancer therapy.
J.Biol.Chem., 285, 2010
|
|
4UQH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
5NNW
 
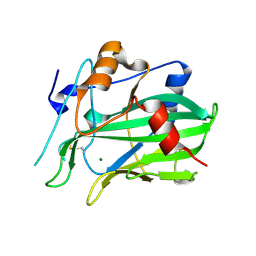 | | NLPPya in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
5NO9
 
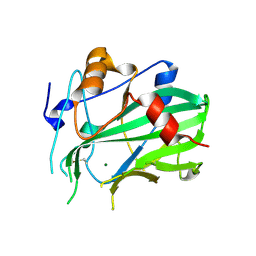 | | NLPPya in complex with mannosamine | | Descriptor: | 2-amino-2-deoxy-alpha-D-mannopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
8E1X
 
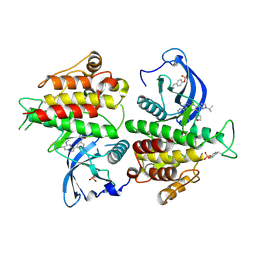 | | FGFR2 kinase domain in complex with a Pyrazolo[1,5-a]pyrimidine analog (Compound 29) | | Descriptor: | (5M)-N-methyl-5-{(6M,8S)-5-{[(3S)-oxolan-3-yl]amino}-6-[1-(propan-2-yl)-1H-pyrazol-3-yl]pyrazolo[1,5-a]pyrimidin-3-yl}pyridine-3-carboxamide, Fibroblast growth factor receptor 2 | | Authors: | Lei, H.-T, Epling, L.B, Deller, M.C. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Wild-Type and Gatekeeper Mutant Fibroblast Growth Factor Receptor (FGFR) 2/3.
J.Med.Chem., 65, 2022
|
|
8W41
 
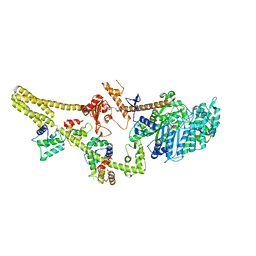 | |
5FCS
 
 | | Diabody | | Descriptor: | Diabody, SULFATE ION | | Authors: | Mosyak, L, Root, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-14 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Development of PF-06671008, a Highly Potent Anti-P-cadherin/Anti-CD3 Bispecific DART Molecule with Extended Half-Life for the Treatment of Cancer.
Antibodies, 5, 2016
|
|
2WEI
 
 | | Crystal structure of the kinase domain of Cryptosporidium parvum calcium dependent protein kinase in complex with 3-MB-PP1 | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALMODULIN-DOMAIN PROTEIN KINASE 1, PUTATIVE | | Authors: | Roos, A.K, King, O, Chaikuad, A, Zhang, C, Shokat, K.M, Wernimont, A.K, Artz, J.D, Lin, L, MacKenzie, F.I, Finerty, P.J, Vedadi, M, Schapira, M, Indarte, M, Kozieradzki, I, Pike, A.C.W, Fedorov, O, Doyle, D, Muniz, J, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Heightman, T, Hui, R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Cryptosporidium Parvum Kinome.
Bmc Genomics, 12, 2011
|
|
8HLO
 
 | | Crystal structure of ASAP1-SH3 and MICAL1-PRM complex | | Descriptor: | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1, Proline rich motif from MICAL1, ... | | Authors: | Niu, F, Wei, Z, Yu, C. | | Deposit date: | 2022-11-30 | | Release date: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | Crystal Structure of the SH3 Domain of ASAP1 in Complex with the Proline Rich Motif (PRM) of MICAL1 Reveals a Unique SH3/PRM Interaction Mode.
Int J Mol Sci, 24, 2023
|
|
1XJD
 
 | |
5CBE
 
 | | E10 in complex with CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, C-X-C motif chemokine 13, E10 heavy chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
5C6W
 
 | | anti-CXCL13 scFv - E10 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
5C2B
 
 | | anti-CXCL13 parental scFv - 3B4 | | Descriptor: | CHLORIDE ION, scFv 3B4 | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-15 | | Release date: | 2015-11-04 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.4049 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
5CBA
 
 | | 3B4 in complex with CXCL13 - 3B4-CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, 3b4 heavy chain, 3b4 light chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
3FXY
 
 | | Acidic Mammalian Chinase, Catalytic Domain | | Descriptor: | Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
3FY1
 
 | | The Acidic Mammalian Chitinase catalytic domain in complex with methylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
3B2Z
 
 | | Crystal Structure of ADAMTS4 (apo form) | | Descriptor: | ADAMTS-4, CALCIUM ION, ZINC ION | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
2RJQ
 
 | | Crystal structure of ADAMTS5 with inhibitor bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, ADAMTS-5, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
2RJP
 
 | | Crystal structure of ADAMTS4 with inhibitor bound | | Descriptor: | ADAMTS-4, CALCIUM ION, N-({4'-[(4-isobutyrylphenoxy)methyl]biphenyl-4-yl}sulfonyl)-D-valine, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | Descriptor: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | Authors: | Silvaggi, N.R, Allen, K.N. | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
6DNA
 
 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6DNB
 
 | | Crystal structure of T110A:S256A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, GLYCEROL, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
