5ZRY
 
 | | Crystal Structure of EphA6/Odin Complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ankyrin repeat and SAM domain-containing protein 1A,Ephrin type-A receptor 6, ... | | 著者 | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5ZRZ
 
 | | Crystal Structure of EphA5/SAMD5 Complex | | 分子名称: | Ephrin type-A receptor 5, Sterile alpha motif domain-containing protein 5 | | 著者 | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | 分子名称: | Pre-mRNA-splicing factor CEF1 | | 著者 | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | 登録日 | 2019-04-14 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | 著者 | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | 登録日 | 2011-07-28 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | 著者 | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | 登録日 | 2011-07-26 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | 著者 | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | 登録日 | 2011-07-28 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
5E5A
 
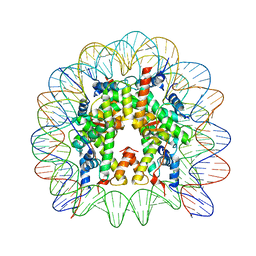 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | 分子名称: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | 著者 | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | 登録日 | 2015-10-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.809 Å) | | 主引用文献 | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
4P7O
 
 | | Structure of Escherichia coli PgaB C-terminal domain, P1 crystal form | | 分子名称: | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | 著者 | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | 登録日 | 2014-03-27 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7Q
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | 著者 | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | 登録日 | 2014-03-27 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7R
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | 著者 | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | 登録日 | 2014-03-27 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7N
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | 著者 | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | 登録日 | 2014-03-27 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P7L
 
 | | Structure of Escherichia coli PgaB C-terminal domain, P212121 crystal form | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | 著者 | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | 登録日 | 2014-03-27 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4JJN
 
 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | 分子名称: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | 著者 | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | 登録日 | 2013-03-08 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5Y36
 
 | | Cryo-EM structure of SpCas9-sgRNA-DNA ternary complex | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, complementary DNA strand, ... | | 著者 | Huang, Q, Li, G, Huai, C. | | 登録日 | 2017-07-27 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | Structural insights into DNA cleavage activation of CRISPR-Cas9 system
Nat Commun, 8, 2017
|
|
7EOZ
 
 | |
7BQA
 
 | | Crystal structure of ASFV p35 | | 分子名称: | 60 kDa polyprotein | | 著者 | Li, G.B, Fu, D, Chen, C, Guo, Y. | | 登録日 | 2020-03-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2021-05-05 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
6KZI
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine derivatives | | 分子名称: | 4-(2-chloro-10H-phenoxazin-10-yl)-N,N-diethylbutan-1-amine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | 登録日 | 2019-09-24 | | 公開日 | 2020-03-04 | | 最終更新日 | 2021-03-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6LJ9
 
 | |
6LJB
 
 | |
6L15
 
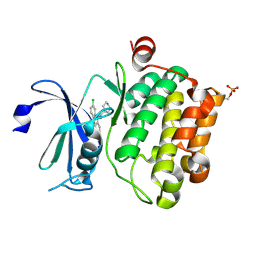 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L12
 
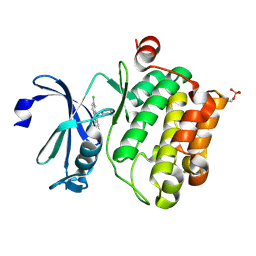 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L16
 
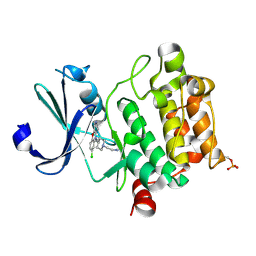 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L11
 
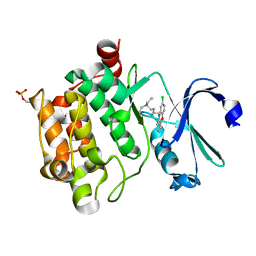 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L17
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-02 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L13
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | 分子名称: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | 著者 | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | 登録日 | 2019-09-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
