8F0Q
 
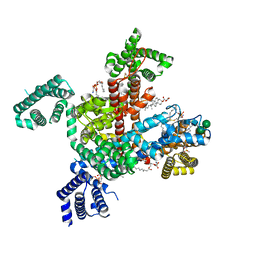 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-cyclopropyl-4-({1-[(1S)-1-(3,5-dichlorophenyl)ethyl]piperidin-4-yl}methoxy)-2-fluoro-N-(methanesulfonyl)benzamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0S
 
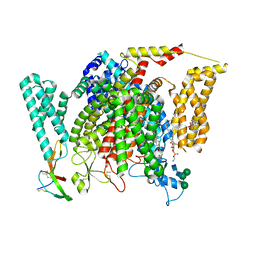 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0P
 
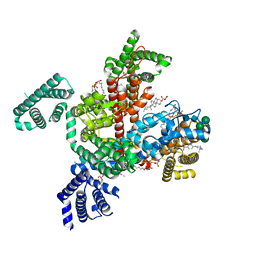 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0R
 
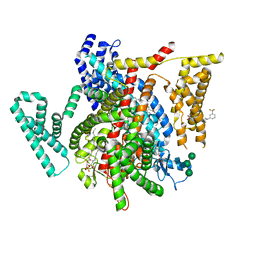 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
7SX3
 
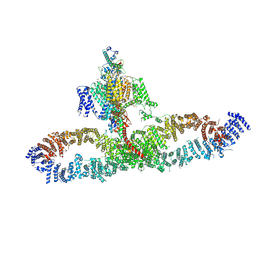 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 1/2 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-29 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
7SX4
 
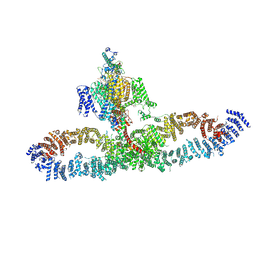 | | Human NALCN-FAM155A-UNC79-UNC80 channelosome with CaM bound, conformation 2/2 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Weidling, C, Chakouri, N, Noland, C.L, Schott, K, Chang, T, Tam, C, Patel, N, Arthur, C.P, Leitner, A, Ben-Johny, M, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-29 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural architecture of the human NALCN channelosome.
Nature, 603, 2022
|
|
6XIW
 
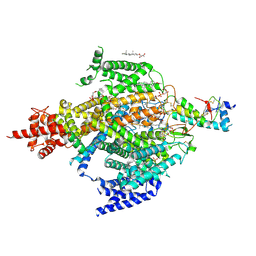 | | Cryo-EM structure of the sodium leak channel NALCN-FAM155A complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Chua, H.C, Noland, C.L, Weidling, C, Clairfeuille, T, Bahlke, O.O, Ameen, A.O, Li, Z.R, Arthur, C.P, Ciferri, C, Pless, S.A, Payandeh, J. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human sodium leak channel NALCN.
Nature, 587, 2020
|
|
7LBE
 
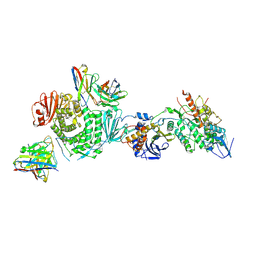 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7T4Q
 
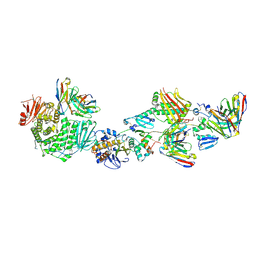 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4S
 
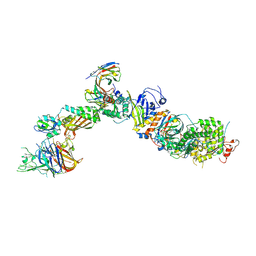 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
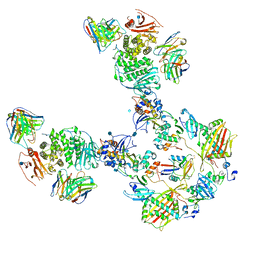 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7LBG
 
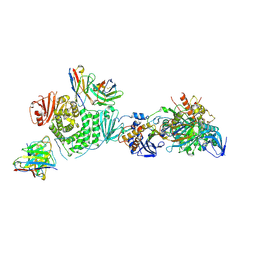 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBF
 
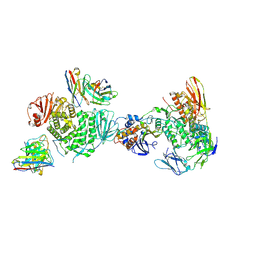 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
5OQR
 
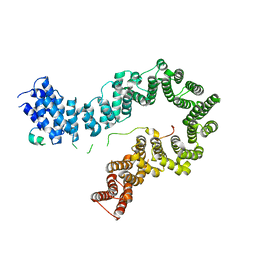 | |
5OQP
 
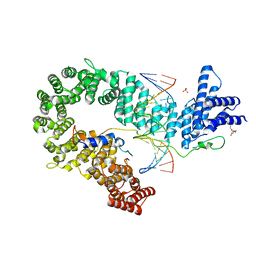 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form I) | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, DNA (5'-D(*GP*AP*TP*GP*TP*GP*TP*AP*GP*CP*TP*AP*CP*AP*CP*AP*TP*C)-3'), ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
5OQO
 
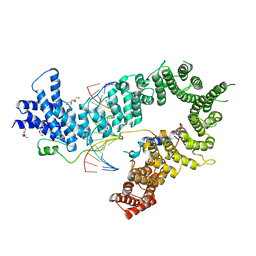 | | Crystal structure of the S. cerevisiae condensin Ycg1-Brn1 subcomplex bound to DNA (crystal form II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Condensin complex subunit 2, Condensin complex subunit 3, ... | | Authors: | Kschonsak, M, Hassler, M, Haering, C.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes.
Cell, 171, 2017
|
|
5OQN
 
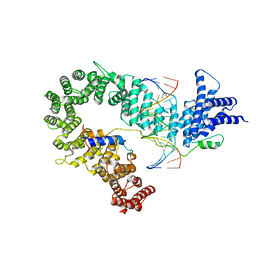 | |
5OQQ
 
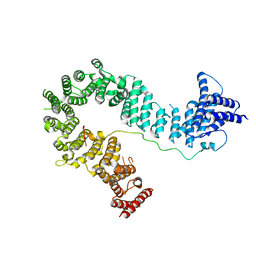 | |
6ZG0
 
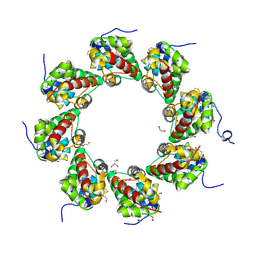 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7ANW
 
 | | hSARM1 NAD+ complex | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Mim, C, Isupov, M.N, Zalk, R, Dessau, M, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-10-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
8AXV
 
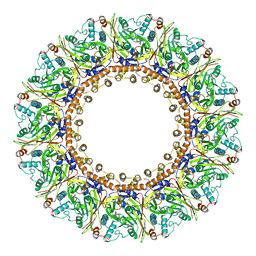 | | Structure of an open form of CHIKV nsP1 capping pores | | Descriptor: | 2-amino-7-methyl-1,7-dihydro-6H-purin-6-one, ZINC ION, mRNA-capping enzyme nsP1 | | Authors: | Reguera, J, Jones, R, Hons, M. | | Deposit date: | 2022-09-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis and dynamics of Chikungunya alphavirus RNA capping by nsP1 capping pores.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8AOX
 
 | |
8AOW
 
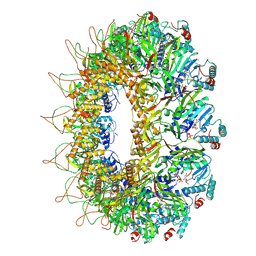 | | CryoEM structure of the Chikungunya virus nsP1 capping pores in complex with m7GTP and SAH ligands | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Jones, R, Hons, M, Reguera, J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis and dynamics of Chikungunya alphavirus RNA capping by nsP1 capping pores.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
