3MFC
 
 | | Computationally designed end0-1,4-beta,xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF6
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3NTB
 
 | | Structure of 6-methylthio naproxen analog bound to mCOX-2. | | Descriptor: | (2S)-2-[6-(methylsulfanyl)naphthalen-2-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NT1
 
 | | High resolution structure of naproxen:COX-2 complex. | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3R3E
 
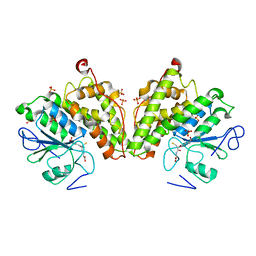 | | The glutathione bound structure of YqjG, a glutathione transferase homolog from Escherichia coli K-12 | | Descriptor: | GLUTATHIONE, SULFATE ION, Uncharacterized protein yqjG | | Authors: | Branch, M.C, Cook, P.D, Harp, J.M, Armstrong, R.N. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure Analysis of yqjG from Escherichia Coli K-12
To be Published
|
|
1MUF
 
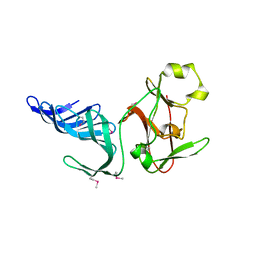 | | Structure of histone H3 K4-specific methyltransferase SET7/9 | | Descriptor: | SET9 | | Authors: | Jacobs, S.A, Harp, J.M, Devarakonda, S, Kim, Y, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The active site of the SET domain is constructed on a knot
Nat.Struct.Biol., 9, 2002
|
|
1OT7
 
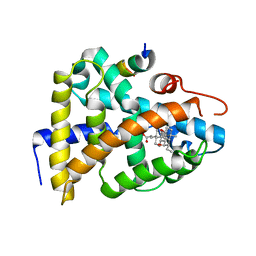 | | Structural Basis for 3-deoxy-CDCA Binding and Activation of FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile Acid Receptor, ISO-URSODEOXYCHOLIC ACID, ... | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OSV
 
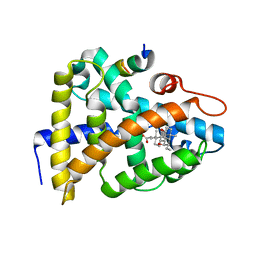 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1QCN
 
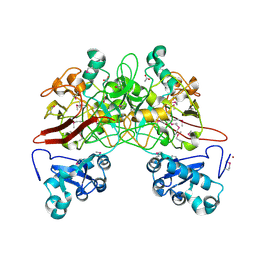 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE | | Descriptor: | ACETATE ION, CALCIUM ION, FUMARYLACETOACETATE HYDROLASE, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-14 | | Release date: | 2000-06-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1QQJ
 
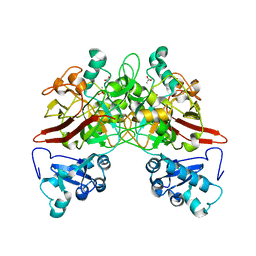 | | CRYSTAL STRUCTURE OF MOUSE FUMARYLACETOACETATE HYDROLASE REFINED AT 1.55 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1R0O
 
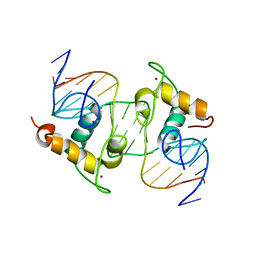 | | Crystal Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex | | Descriptor: | Ecdysone Response Element, Ecdysone receptor, Ultraspiracle protein, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex
Embo J., 22, 2003
|
|
1QCO
 
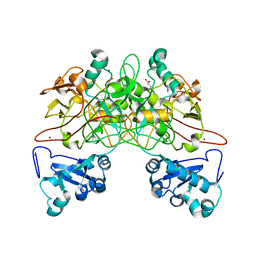 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH FUMARATE AND ACETOACETATE | | Descriptor: | ACETOACETIC ACID, CALCIUM ION, FUMARIC ACID, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-17 | | Release date: | 2000-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1R0N
 
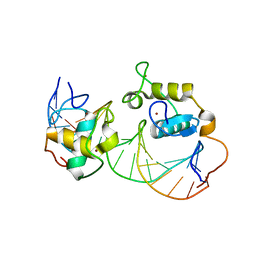 | | Crystal Structure of Heterodimeric Ecdsyone receptor DNA binding complex | | Descriptor: | Ecdsyone Response Element, Ecdysone Response Element, Ecdysone receptor, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the heterodimeric Ecdysone Receptor DNA-binding complex
Embo J., 22, 2003
|
|
3TDM
 
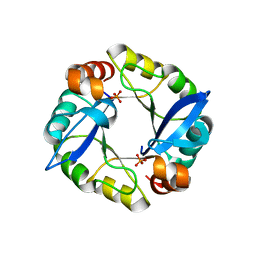 | | Computationally designed TIM-barrel protein, HalfFLR | | Descriptor: | Computationally designed two-fold symmetric TIM-barrel protein, FLR (half molecule), PHOSPHATE ION | | Authors: | Harp, J.M, Fortenberry, C, Bowman, E, Profitt, W, Dorr, B, Mizoue, L. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploring symmetry as an avenue to the computational design of large protein domains.
J.Am.Chem.Soc., 133, 2011
|
|
3TDN
 
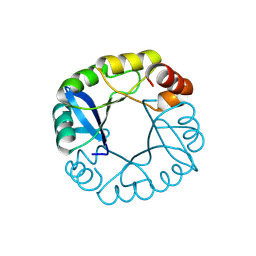 | | Computationally designed two-fold symmetric Tim-barrel protein, FLR | | Descriptor: | FLR SYMMETRIC ALPHA-BETA TIM BARREL | | Authors: | Harp, J.M, Fortenberry, C, Bowman, E, Profitt, W, Dorr, B, Mizoue, L, Meiler, J. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring symmetry as an avenue to the computational design of large protein domains.
J.Am.Chem.Soc., 133, 2011
|
|
5DER
 
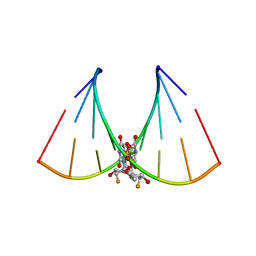 | | RNA oligonucleotide containing (R)-C5'-ME-2'F U | | Descriptor: | RNA oligonucleotide containing (R)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5D8T
 
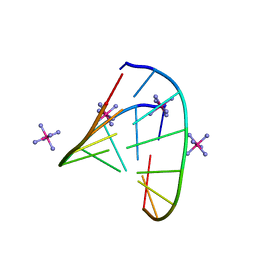 | | RNA octamer containing (S)-5' methyl, 2'-F U. | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing (S)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5DEK
 
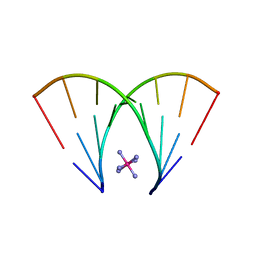 | | RNA octamer containing dT | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing dT | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
5V2H
 
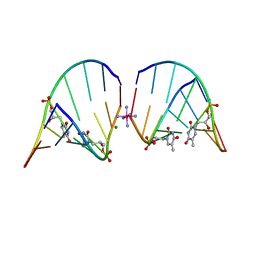 | | RNA octamer containing glycol nucleic acid, SgnT | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, RNA (5'-R(*(CBV)P*GP*AP*AP*(ZTH)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (1.080013 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
5VR4
 
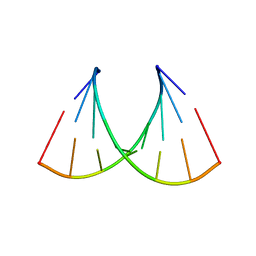 | | RNA octamer containing 2'-F-4'-OMe U. | | Descriptor: | COBALT TETRAAMMINE ION, RNA (5'-R(*CP*GP*AP*AP*(UMO)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing.
J. Am. Chem. Soc., 139, 2017
|
|
3C8E
 
 | | Crystal Structure Analysis of yghU from E. Coli | | Descriptor: | GLUTATHIONE, yghU, glutathione S-transferase homologue | | Authors: | Harp, J, Ladner, J.E, Schaab, M.R, Stourman, N.V, Armstrong, R.N. | | Deposit date: | 2008-02-11 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Function of YghU, a Nu-Class Glutathione Transferase Related to YfcG from Escherichia coli.
Biochemistry, 50, 2011
|
|
5HFK
 
 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
3G1Q
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in ligand free state | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha-demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Harp, J, Wawrzak, Z, Waterman, M.R, Park, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections.
J.Biol.Chem., 285, 2010
|
|
3GW9
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei bound to an inhibitor N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxaziazol-2-yl)benzamide | | Descriptor: | N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14ALPHA-DEMETHYLASE | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Harp, J, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections.
J.Biol.Chem., 285, 2010
|
|
2GLK
 
 | | High-resolution study of D-Xylose isomerase, 0.94A resolution. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Carrell, H.L, Hanson, B.L, Harp, J.M, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
