4AY9
 
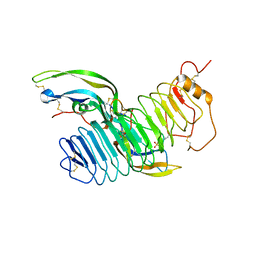 | | Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLLICLE-STIMULATING HORMONE RECEPTOR, FOLLITROPIN SUBUNIT BETA, ... | | Authors: | Jiang, X, Liu, H, Chen, X, He, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Follicle-Stimulating Hormone in Complex with the Entire Ectodomain of its Receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HKA
 
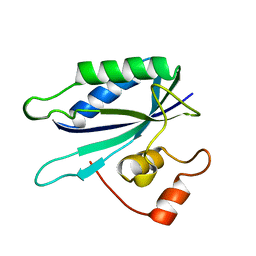 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
2RFU
 
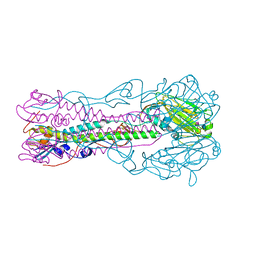 | | Crystal structure of influenza B virus hemagglutinin in complex with LSTc receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Influenza B hemagglutinin (HA), ... | | Authors: | Wang, Q, Tian, X, Chen, X, Ma, J. | | Deposit date: | 2007-10-01 | | Release date: | 2008-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for receptor specificity of influenza B virus hemagglutinin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RFT
 
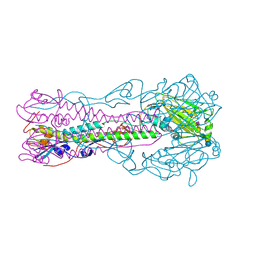 | | Crystal structure of influenza B virus hemagglutinin in complex with LSTa receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Influenza B hemagglutinin (HA), ... | | Authors: | Wang, Q, Tian, X, Chen, X, Ma, J. | | Deposit date: | 2007-10-01 | | Release date: | 2008-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for receptor specificity of influenza B virus hemagglutinin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1WWN
 
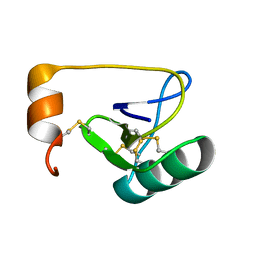 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
5GWZ
 
 | | The structure of Porcine epidemic diarrhea virus main protease in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, PEDV main protease | | Authors: | Wang, F, Chen, C, Yang, K, Liu, X, Liu, H, Xu, Y, Chen, X, Liu, X, Cai, Y, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
J. Med. Chem., 60, 2017
|
|
4GHQ
 
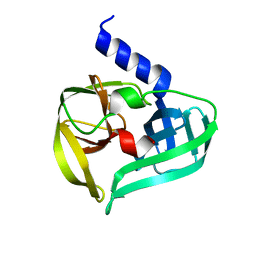 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VCB
 
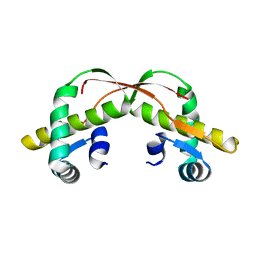 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3VC8
 
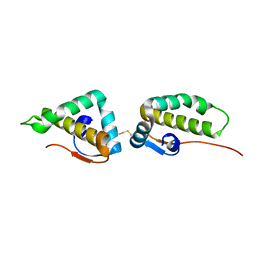 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
1YVL
 
 | | Structure of Unphosphorylated STAT1 | | Descriptor: | 5-residue peptide, GOLD ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Mao, X, Ren, Z, Parker, G.N, Sondermann, H, Pastorello, M.A, Wang, W, McMurray, J.S, Demeler, B, Darnell Jr, J.E, Chen, X. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural bases of unphosphorylated STAT1 association and receptor binding.
Mol.Cell, 17, 2005
|
|
7QZQ
 
 | | Crystal structure of the kelch domain of human KBTBD12 | | Descriptor: | 1,2-ETHANEDIOL, Kelch repeat and BTB domain-containing protein 12, SODIUM ION | | Authors: | Manning, C.E, Chen, Z, Chen, X, Bradshaw, W.J, Bakshi, S, Mckinley, G, Chalk, R, Burgess-Brown, N, von Delft, F, Bullock, A.N. | | Deposit date: | 2022-01-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the kelch domain of human KBTBD12
To Be Published
|
|
6CIL
 
 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | Descriptor: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIK
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-intact/nicked 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 12RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIM
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-nicked/intact 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 23RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
1FIO
 
 | | CRYSTAL STRUCTURE OF YEAST T-SNARE PROTEIN SSO1 | | Descriptor: | SSO1 PROTEIN, ZINC ION | | Authors: | Munson, M, Chen, X, Cocina, A.E, Schultz, S.M, Hughson, F.M. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly.
Nat.Struct.Biol., 7, 2000
|
|
7RR5
 
 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
7RSL
 
 | | Seipin forms a flexible cage at lipid droplet formation sites | | Descriptor: | Seipin | | Authors: | Arlt, H, Sui, X, Folger, B, Adams, C, Chen, X, Remme, R, Hamprecht, F.A, DiMaio, F, Liao, M, Goodman, J.M, Farese Jr, R.V, Walther, T.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Seipin forms a flexible cage at lipid droplet formation sites.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1GH2
 
 | | Crystal structure of the catalytic domain of a new human thioredoxin-like protein | | Descriptor: | THIOREDOXIN-LIKE PROTEIN | | Authors: | Jin, J, Chen, X, Guo, Q, Yuan, J, Qiang, B, Rao, Z. | | Deposit date: | 2000-11-01 | | Release date: | 2001-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the catalytic domain of a human thioredoxin-like protein.
Eur.J.Biochem., 269, 2002
|
|
1IK1
 
 | | Solution Structure of an RNA Hairpin from HRV-14 | | Descriptor: | 5'-R(*GP*GP*UP*AP*CP*UP*AP*UP*GP*UP*AP*CP*CP*A)-3' | | Authors: | Huang, H, Alexandrov, A, Chen, X, Barnes III, T.W, Zhang, H, Dutta, K, Pascal, S.M. | | Deposit date: | 2001-05-01 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin from HRV-14.
Biochemistry, 40, 2001
|
|
8Z1E
 
 | | A homotrimeric GPCR architecture of the human cytomegalovirus (UL78) revealed by cryo-EM | | Descriptor: | Uncharacterized protein UL78 | | Authors: | Chen, Y, Li, Y, Zhou, Q, Cong, Z, Lin, S, Yan, J, Chen, X, Yang, D, Ying, T, Wang, M.-W. | | Deposit date: | 2024-04-11 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A homotrimeric GPCR architecture of the human cytomegalovirus revealed by cryo-EM
To Be Published
|
|
2QVU
 
 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Mg2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
2QVV
 
 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Zn2+, I(T)-state | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, PHOSPHATE ION, ... | | Authors: | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
1C8J
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F) | | Descriptor: | CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Y, Jiang, F, Guo, Q, Chen, X, Jin, J, Sun, Y, Rao, Z. | | Deposit date: | 2000-05-31 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450cam mutant (F87W/Y96F)
To be Published
|
|
