7SIM
 
 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4KYK
 
 | | Crystal structure of mouse glyoxalase I complexed with indomethacin | | Descriptor: | INDOMETHACIN, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
4KYH
 
 | | Crystal structure of mouse glyoxalase I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
7F8L
 
 | |
6KEE
 
 | | Crystal structure of BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12154651 Å) | | Cite: | Crystal structure of BRD4 Bromodomain1 with an inhibitor
To be published
|
|
5Z90
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 1-ethyl-6-pyrrolidin-1-ylsulfonyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5Z8G
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 1-ethyl-6-[(3R)-3-oxidanylpiperidin-1-yl]sulfonyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-01-31 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5Z8R
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 1-ethyl-6-[(3S)-3-oxidanylpiperidin-1-yl]sulfonyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5Z9K
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 1-ethyl-6-[(2R)-2-(hydroxymethyl)pyrrolidin-1-yl]sulfonyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5Z8Z
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 6-[(3S)-3-azanylpiperidin-1-yl]sulfonyl-1-ethyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
8KDB
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
8KDC
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
7CI3
 
 | |
4NFT
 
 | | Crystal structure of human lnkH2B-h2A.Z-Anp32e | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member E, Histone H2B type 2-E, Histone H2A.Z | | Authors: | Shan, S, Pan, L, Mao, Z, Wang, W, Sun, J, Dong, Q, Liang, X, Ding, X, Chen, S, Dai, L, Zhang, Z, Zhu, B, Zhou, Z. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z
Cell Res., 24, 2014
|
|
7DWS
 
 | |
4GQG
 
 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
4I5X
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
3R8W
 
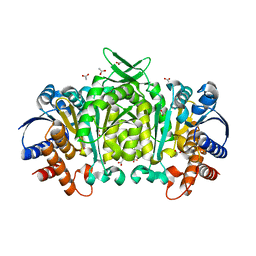 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | Authors: | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
6KEG
 
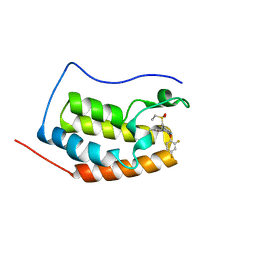 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(ethylsulfonylmethyl)pyridin-3-yl]-8-methyl-4H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KEF
 
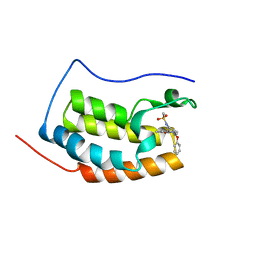 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(8-methyl-1-oxidanylidene-2H-pyrrolo[1,2-a]pyrazin-6-yl)-4-phenoxy-phenyl]methanesulfonamide | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44467545 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KED
 
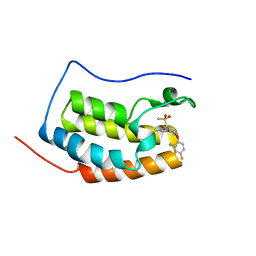 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-d][1,2,4]triazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.548447 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
5GOV
 
 | | Crystal Structure of MCR-1, a phosphoethanolamine transferase, extracellular domain | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hu, M, Guo, J, Chen, S, Hao, Q. | | Deposit date: | 2016-07-29 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for Colistin Resistance.
Sci Rep, 6, 2016
|
|
3EBJ
 
 | | Crystal structure of an avian influenza virus protein | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein | | Authors: | Yuan, P, Bartlam, M, Lou, Z, Chen, S, Rao, Z, Liu, Y. | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site
Nature, 458, 2009
|
|
