5FM8
 
 | |
8AJR
 
 | |
8ALQ
 
 | |
6P06
 
 | |
6VRV
 
 | | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model | | Descriptor: | 4-chloro-1-{(1S)-1-[(3S)-3-fluoropyrrolidin-3-yl]ethyl}-3-methyl-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Barberis, C.E, Batchelor, J.D, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6VRU
 
 | | PIM-inhibitor complex 1 | | Descriptor: | 3,4-dichloro-2-cyclopropyl-1-[(piperidin-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Barberis, C.E, Batchelor, J.D, Mechin, I, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WAQ
 
 | |
4JID
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | Descriptor: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
6PLF
 
 | | Crystal structure of human PHGDH complexed with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-{(1S)-1-[(5-chloro-6-{[(5S)-2-oxo-1,3-oxazolidin-5-yl]methoxy}-1H-indole-2-carbonyl)amino]-2-hydroxyethyl}benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4JKM
 
 | |
7OSL
 
 | |
7P6Y
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH compound 5ef | | Descriptor: | 4-benzoyl-N-(2-(2-(2-((2-(1,5-dimethyl-6-oxo-1,6-dihydropyridin-3-yl)-1-((tetrahydro-2H-pyran-4-yl)methyl)-1H-benzo[d]imidazol-6-yl)(methyl)amino)ethoxy)ethoxy)ethyl)-N-(2-oxo-2-((2-(2-(prop-2-yn-1-yloxy)ethoxy)ethyl)amino)ethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2021-07-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | One-Step Synthesis of Photoaffinity Probes for Live-Cell MS-Based Proteomics.
Chemistry, 27, 2021
|
|
7P6V
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH compound 3ag | | Descriptor: | Bromodomain-containing protein 4, ethyl 2-(2-(4-azido-N-((2-(1,5-dimethyl-6-oxo-1,6-dihydropyridin-3-yl)-1-((tetrahydro-2H-pyran-4-yl)methyl)-1H-benzo[d]imidazol-6-yl)methyl)-2,3,5,6-tetrafluorobenzamido)acetamido)acetate | | Authors: | Chung, C. | | Deposit date: | 2021-07-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | One-Step Synthesis of Photoaffinity Probes for Live-Cell MS-Based Proteomics.
Chemistry, 27, 2021
|
|
7P6W
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH compound 3bg | | Descriptor: | Bromodomain-containing protein 4, ethyl 2-(2-(4-azido-N-((2-(1,5-dimethyl-6-oxo-1,6-dihydropyridin-3-yl)-1-((tetrahydro-2H-pyran-4-yl)methyl)-1H-benzo[d]imidazol-6-yl)methyl)benzamido)acetamido)acetate | | Authors: | Chung, C. | | Deposit date: | 2021-07-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | One-Step Synthesis of Photoaffinity Probes for Live-Cell MS-Based Proteomics.
Chemistry, 27, 2021
|
|
7U8E
 
 | |
7OVD
 
 | | Human soluble adenylyl cyclase in complex with the inhibitor TDI10229 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-6-[1,5-dimethyl-4-(phenylmethyl)pyrazol-3-yl]pyrimidin-2-amine, ACETATE ION, ... | | Authors: | Steegborn, C, Quast, J. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of TDI-10229: A Potent and Orally Bioavailable Inhibitor of Soluble Adenylyl Cyclase (sAC, ADCY10).
Acs Med.Chem.Lett., 12, 2021
|
|
6VL3
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986436 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-22 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7OMB
 
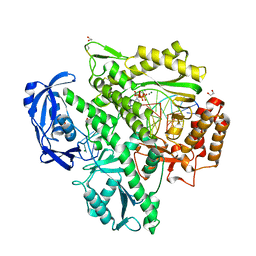 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OMG
 
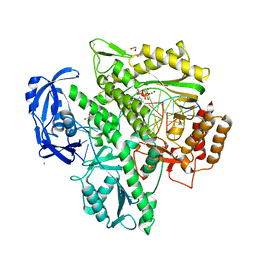 | | Crystal structure of KOD DNA Polymerase in a ternary complex with an Uracil containing template | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OM3
 
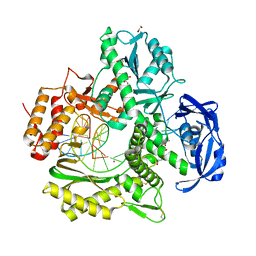 | | Crystal structure of KOD DNA Polymerase in a binary complex with Hypoxanthine containing template | | Descriptor: | 1,2-ETHANEDIOL, 21nt Template, BROMIDE ION, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
5FM4
 
 | |
1PPG
 
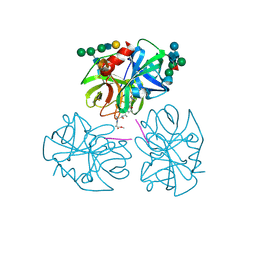 | | The refined 2.3 angstroms crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor | | Descriptor: | HUMAN LEUKOCYTE ELASTASE, MEO-SUCCINYL-ALA-ALA-PRO-VAL CHLOROMETHYLKETONE, alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bode, W, Wei, A-Z. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The refined 2.3 A crystal structure of human leukocyte elastase in a complex with a valine chloromethyl ketone inhibitor.
FEBS Lett., 234, 1988
|
|
6BTP
 
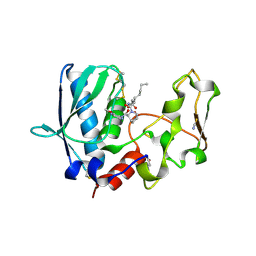 | | BMP1 complexed with a hydroxamate | | Descriptor: | (1R,3S,4S)-2-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]heptanoyl}-N-(3-methoxypyrazin-2-yl)-2-azabicyclo[2.2.1]heptane-3-carboxamide, Bone morphogenetic protein 1, THIOCYANATE ION, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
7UP4
 
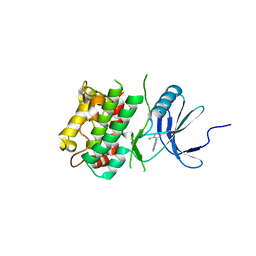 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 20 (co-crystal) | | Descriptor: | (5M)-5-(2,5-dichloropyrimidin-4-yl)-5H-pyrrolo[3,2-d]pyrimidine, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP6
 
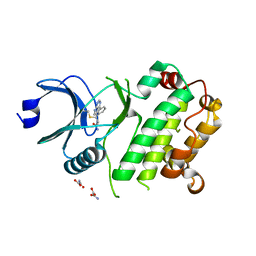 | | Crystal structure of C-terminal domain of MSK1 in complex with in covalently bound literature RSK2 inhibitor pyrrolopyrimidine cyanoacrylamide compound 25 (co-crystal) | | Descriptor: | (E)-3-(3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl)-2-cyanoacrylamide bound form, OXAMIC ACID, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
