2I4J
 
 | |
7M3I
 
 | |
2KW6
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 1 (CDK2-associated protein 1; oral cancer suppressor Deleted in oral cancer 1, DOC-1) from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3057H | | Descriptor: | Cyclin-dependent kinase 2-associated protein 1 | | Authors: | Ertekin, A, Aramini, J.M, Rossi, P, Lee, A.B, Jiang, M, Ciccosanti, C.T, Xiao, R, Swapna, G.V.T, Rost, B, Everett, J.K, Acton, T.B, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Human cyclin-dependent kinase 2-associated protein 1 (CDK2AP1) is dimeric in its disulfide-reduced state, with natively disordered N-terminal region.
J.Biol.Chem., 287, 2012
|
|
2I4Z
 
 | |
2LPR
 
 | |
3EQB
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | 2-Alkylamino- and alkoxy-substituted 2-amino-1,3,4-oxadiazoles-O-Alkyl benzohydroxamate esters replacements retain the desired inhibition and selectivity against MEK (MAP ERK kinase).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3GN4
 
 | | Myosin lever arm | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Mukherjea, M, Llinas, P, Kim, H, Travaglia, M, Safer, D, Zong, A.B, Menetrey, J, Franzini-Armstrong, C, Selvin, P.R, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach
Mol.Cell, 35, 2009
|
|
8FNZ
 
 | | Acetylated tau repeat 1 and 2 fragment (AcR1R2) | | Descriptor: | Microtubule-associated protein tau, acetylated repeat 1 and 2 fragment | | Authors: | Li, L, Nguyen, A.B, Mullapudi, V, Joachimiak, L. | | Deposit date: | 2022-12-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Disease-associated patterns of acetylation stabilize tau fibril formation.
Structure, 31, 2023
|
|
7JMW
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
1IHV
 
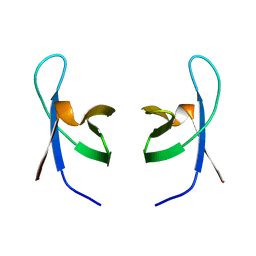 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1IHW
 
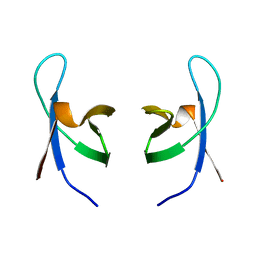 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1LKI
 
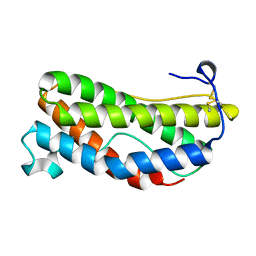 | | THE CRYSTAL STRUCTURE AND BIOLOGICAL FUNCTION OF LEUKEMIA INHIBITORY FACTOR: IMPLICATIONS FOR RECEPTOR BINDING | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Grey, L.M, Staunton, D, Stuart, D.I, Heath, J.K, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding.
Cell(Cambridge,Mass.), 77, 1994
|
|
1IQC
 
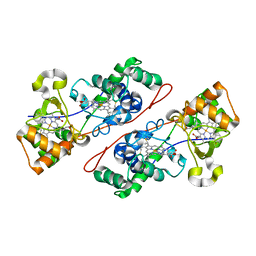 | | Crystal structure of Di-Heme Peroxidase from Nitrosomonas europaea | | Descriptor: | CALCIUM ION, GLYCEROL, HEME C, ... | | Authors: | Shimizu, H. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Nitrosomonas europaea cytochrome c peroxidase and the structural basis for ligand switching in bacterial di-heme peroxidases
Biochemistry, 40, 2001
|
|
7LM8
 
 | |
7LM9
 
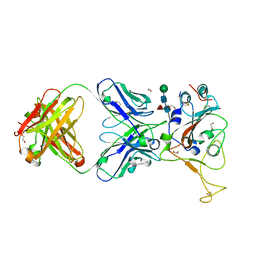 | | Crystal structure of SARS-CoV spike protein receptor-binding domain in complex with a cross-neutralizing antibody CV38-142 Fab isolated from COVID-19 patient | | Descriptor: | 1,2-ETHANEDIOL, CV38-142 Fab heavy chain, CV38-142 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection.
Cell Host Microbe, 29, 2021
|
|
7S0J
 
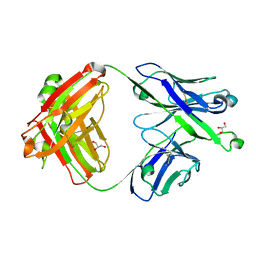 | | Crystal structure of Epstein-Barr virus gH/gL targeting antibody 769B10 | | Descriptor: | 769B10 Fab Heavy chain, 769B10 Fab Light chain, GLYCEROL | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S08
 
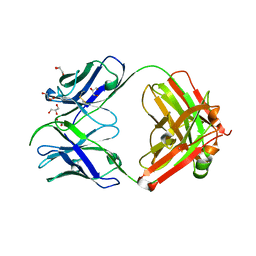 | |
7S1B
 
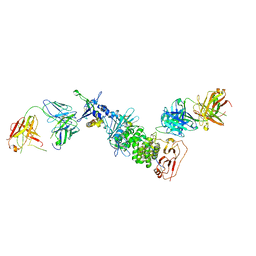 | | Crystal structure of Epstein-Barr virus glycoproteins gH/gL/gp42-peptide in complex with human neutralizing antibodies 769C2 and 770F7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 Fab heavy chain, 769C2 Fab light chain, ... | | Authors: | Chen, W.-H, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S07
 
 | | Crystal structure of Epstein-Barr virus glycoprotein gH/gL/gp42-peptide in complex with human neutralizing antibodies 769B10 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769B10 Fab heavy chain, 769B10 Fab light chain, ... | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S83
 
 | | Crystal structure of SARS CoV-2 Spike Receptor Binding Domain in complex with shark neutralizing VNARs ShAb01 and ShAb02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ShAb01 VNAR, ... | | Authors: | Chen, W.-H, Hajduczki, A, Dooley, H.M, Joyce, M.G. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity.
Nat Commun, 14, 2023
|
|
7TB4
 
 | | Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Antibodies with potent and broad neutralizing activity against antigenically diverse and highly transmissible SARS-CoV-2 variants.
Biorxiv, 2021
|
|
7TE1
 
 | |
7TDZ
 
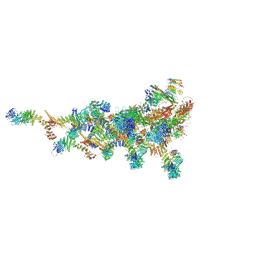 | |
7SRS
 
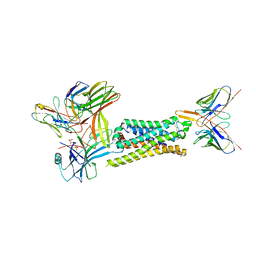 | | 5-HT2B receptor bound to LSD in complex with beta-arrestin1 obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 2B, ... | | Authors: | Barros-Alvarez, X, Cao, C, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
7SRQ
 
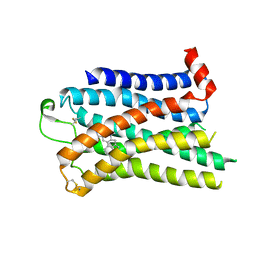 | | 5-HT2B receptor bound to LSD obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 2B | | Authors: | Barros-Alvarez, X, Cao, C, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
