7RRA
 
 | |
5UKQ
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.2 Fab | | Descriptor: | DH522.2 Fab fragment heavy chain, DH522.2 Fab fragment light chain, GLYCEROL | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
3QHD
 
 | |
8ET1
 
 | |
8ET2
 
 | | CryoEM structure of the GSDMB pore | | Descriptor: | Isoform 1 of Gasdermin-B | | Authors: | Wang, C, Ruan, J. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.96 Å) | | Cite: | Structural basis for GSDMB pore formation and its targeting by IpaH7.8.
Nature, 616, 2023
|
|
7ML7
 
 | |
7K99
 
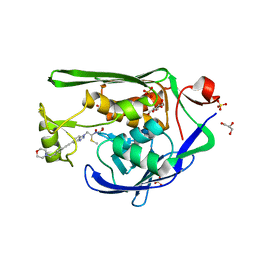 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 | | Descriptor: | (hydroxy{(1S)-1-(methylsulfanyl)-2-[5-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)-1H-benzotriazol-1-yl]ethyl}amino)methanol, GLYCEROL, SULFATE ION, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
3B5H
 
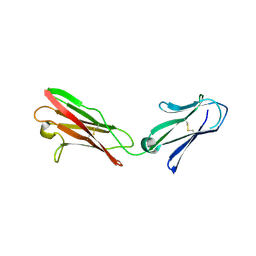 | |
7JTM
 
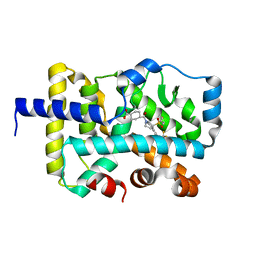 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(6T78-692) IN COMPLEX WITH A TRICYCLIC SULFONE RORGT INVERSE AGONIST | | Descriptor: | Nuclear receptor ROR-gamma, trans-4-[(3aR,9bR)-8-cyano-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]cyclohexane-1-carboxylic acid | | Authors: | Sack, J.S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Tricyclic sulfones as potent, selective and efficacious ROR gamma t inverse agonists - Exploring C6 and C8 SAR using late-stage functionalization.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6W9H
 
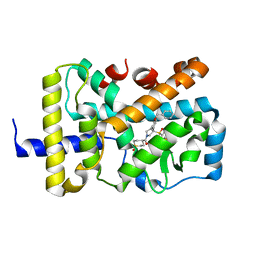 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | 4-{(3R)-3-[4-(benzyloxy)phenyl]-3-[(4-fluorophenyl)sulfonyl]pyrrolidine-1-carbonyl}-1lambda~6~-thiane-1,1-dione, Nuclear receptor ROR-gamma, Steroid receptor coactivator 1 fusion | | Authors: | Sack, J.S. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2YER
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 5-(HYDROXYMETHYL)-8-(1H-PYRROL-2-YL)-2H-[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A, Otterbein, L. | | Deposit date: | 2011-03-30 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6W9I
 
 | |
5KZA
 
 | | Crystal structure of the Rous sarcoma virus matrix protein (aa 2-102). Space group I41 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, virus matrix protein | | Authors: | Kingston, R.L, Chan, J, Vogt, V.M. | | Deposit date: | 2016-07-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cholesterol Promotes Protein Binding by Affecting Membrane Electrostatics and Solvation Properties.
Biophys. J., 113, 2017
|
|
7JWL
 
 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | Descriptor: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
7K88
 
 | | Crystal structure of bovine RPE65 in complex with hexaethylene glycol monooctyl ether | | Descriptor: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, FE (II) ION, Retinoid isomerohydrolase, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2020-09-26 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
J.Med.Chem., 64, 2021
|
|
5T8I
 
 | | PI3Kdelta in complex with the inhibitor GS-9901 | | Descriptor: | 2,4-diamino-6-{[(S)-[5-chloro-8-fluoro-4-oxo-3-(pyridin-3-yl)-3,4-dihydroquinazolin-2-yl](cyclopropyl)methyl]amino}pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A. | | Deposit date: | 2016-09-07 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of GS-9901: A Potent, Selective and Metabolically Stable Inhibitor of PI3Kd
To Be Published
|
|
7K8G
 
 | | Crystal structure of bovine RPE65 in complex with 4-fluoro-MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{4-fluoro-3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, FE (II) ION, PALMITIC ACID, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
J.Med.Chem., 64, 2021
|
|
7K89
 
 | |
3PUI
 
 | | Cocaine Esterase with mutations G4C, S10C | | Descriptor: | CHLORIDE ION, Cocaine esterase, SODIUM ION | | Authors: | Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2010-12-04 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Subunit stabilization and polyethylene glycolation of cocaine esterase improves in vivo residence time.
Mol.Pharmacol., 80, 2011
|
|
6UIK
 
 | |
6UIF
 
 | |
4AIS
 
 | | A complex structure of BtGH84 | | Descriptor: | GLYCEROL, GLYCOLIC ACID, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
7L0E
 
 | |
6XAE
 
 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | Nuclear receptor ROR-gamma, trans-4-{(3aR,9bR)-7-[(2-chloro-6-fluorophenyl)methoxy]-9b-[(4-fluorophenyl)sulfonyl]-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl}cyclohexane-1-carboxylic acid | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
