2I75
 
 | | Crystal Structure of Human Protein Tyrosine Phosphatase N4 (PTPN4) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Ugochukwu, E, Barr, A, Savitsky, P, Burgess, N, Das, S, Turnbull, A, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2H4V
 
 | | Crystal Structure of the Human Tyrosine Receptor Phosphatase Gamma | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Das, S, Eswaran, J, Savitsky, P, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Debreczeni, J, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2J7T
 
 | | Crystal structure of human serine threonine kinase-10 bound to SU11274 | | Descriptor: | (3Z)-N-(3-CHLOROPHENYL)-3-({3,5-DIMETHYL-4-[(4-METHYLPIPERAZIN-1-YL)CARBONYL]-1H-PYRROL-2-YL}METHYLENE)-N-METHYL-2-OXOINDOLINE-5-SULFONAMIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Das, S, Debreczeni, J, Sobott, F, Watt, S, Savitsky, P, Eswaran, J, Turnbull, A.P, Papagrigoriou, E, Ugochukwa, E, Gorrec, F, Umeano, C.C, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2F57
 
 | | Crystal Structure Of The Human P21-Activated Kinase 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE, Serine/threonine-protein kinase PAK 7 | | Authors: | Eswaran, J, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Bray, J, Das, S, Savitsky, P, Smee, C, Burgess, N, Fedorov, O, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs.
Structure, 15, 2007
|
|
8DDL
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Apo Structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ORF1a polyprotein, ... | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
8DD6
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant in Complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, DIMETHYL SULFOXIDE, ORF1a polyprotein | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-17 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
2C30
 
 | | Crystal Structure Of The Human P21-Activated Kinase 6 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN KINASE PAK 6 | | Authors: | Filippakopoulos, P, Berridge, G, Bray, J, Burgess, N, Colebrook, S, Das, S, Eswaran, J, Gileadi, O, Papagrigoriou, E, Savitsky, P, Smee, C, Turnbull, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Knapp, S. | | Deposit date: | 2005-10-02 | | Release date: | 2006-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the P21-Activated Kinases Pak4, Pak5, and Pak6 Reveal Catalytic Domain Plasticity of Active Group II Paks.
Structure, 15, 2007
|
|
2AHS
 
 | | Crystal Structure of the Catalytic Domain of Human Tyrosine Receptor Phosphatase Beta | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta, ... | | Authors: | Ugochukwu, E, Eswaran, J, Barr, A, Gileadi, O, Sobott, F, Burgess, N, Ball, L, Bray, J, von Delft, F, Debreczeni, J, Bunkoczi, G, Turnbull, A, Das, S, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2B49
 
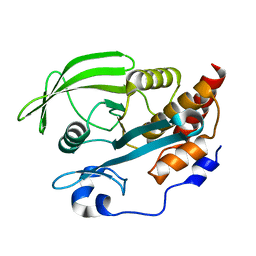 | | Crystal Structure of the Catalytic Domain of Protein Tyrosine Phosphatase, non-receptor Type 3 | | Descriptor: | protein tyrosine phosphatase, non-receptor type 3 | | Authors: | Ugochukwu, E, Arrowsmith, C, Barr, A, Bunkoczi, G, Das, S, Debreczeni, J, Edwards, A, Eswaran, J, Knapp, S, Sundstrom, M, Turnbull, A, von Delft, F, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2A3K
 
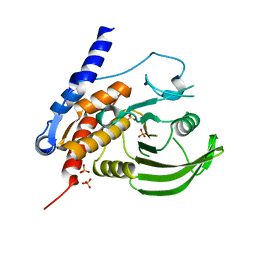 | | Crystal Structure of the Human Protein Tyrosine Phosphatase, PTPN7 (HePTP, Hematopoietic Protein Tyrosine Phosphatase) | | Descriptor: | PHOSPHATE ION, protein tyrosine phosphatase, non-receptor type 7, ... | | Authors: | Barr, A, Turnbull, A.P, Das, S, Eswaran, J, Debreczeni, J.E, Longmann, E, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-24 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
2BZL
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PROTEIN TYROSINE PHOSPHATASE N14 AT 1. 65 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE, ... | | Authors: | Debreczeni, J.E, Barr, A, Eswaran, J, Das, S, Burgess, N, Longman, E, Fedorov, O, Gileadi, O, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2005-08-18 | | Release date: | 2005-09-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Human Protein Tyrosine Phosphatase 14 (Ptpn14) at 1.65-A Resolution.
Proteins, 63, 2006
|
|
2BVA
 
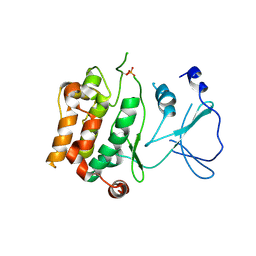 | | Crystal structure of the human P21-activated kinase 4 | | Descriptor: | P21-ACTIVATED KINASE 4 | | Authors: | Debreczeni, J.E, Bunkoczi, G, Eswaran, J, Filippakopoulos, P, Das, S, Fedorov, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S. | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the P21-Activated Kinases Pak4, Pak5, and Pak6 Reveal Catalytic Domain Plasticity of Active Group II Paks.
Structure, 15, 2007
|
|
2CDZ
 
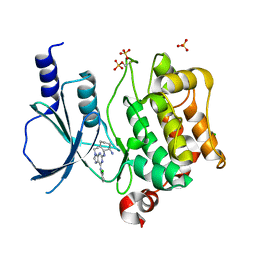 | | CRYSTAL STRUCTURE OF THE HUMAN P21-ACTIVATED KINASE 4 IN COMPLEX WITH CGP74514A | | Descriptor: | CHLORIDE ION, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE, SERINE/THREONINE-PROTEIN KINASE PAK 4, ... | | Authors: | Debreczeni, J.E, Ugochukwu, E, Eswaran, J, Filippakopoulos, P, Das, S, Fedorov, O, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Knapp, S. | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the P21-Activated Kinases Pak4, Pak5, and Pak6 Reveal Catalytic Domain Plasticity of Active Group II Paks.
Structure, 15, 2007
|
|
2C7S
 
 | | Crystal structure of human protein tyrosine phosphatase kappa at 1.95A resolution | | Descriptor: | ACETATE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE KAPPA | | Authors: | Debreczeni, J.E, Ugochukwu, E, Eswaran, J, Barr, A, Das, S, Burgess, N, Gileadi, O, Longman, E, von Delft, F, Knapp, S, Sundstron, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-11-28 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
8ARL
 
 | | Plasmodium vivax PVP01_0000100 TRAg domain | | Descriptor: | Tryptophan-rich antigen | | Authors: | Kundu, P, Deane, J.E, Rayner, J.C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a Plasmodium vivax Tryptophan Rich Antigen domain suggests a lipid binding function for a pan-Plasmodium multi-gene family.
Nat Commun, 14, 2023
|
|
6MIH
 
 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIK
 
 | | Crystal structure of host-guest complex with PP hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIG
 
 | | Crystal structure of host-guest complex with PB hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), Gag-Pol polyprotein | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
8SG6
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Reduced with 20mM TCEP | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Tran, N, McLeod, M.J, Barwell, S, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2023-04-11 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
8SM5
 
 | |
5DSE
 
 | | Crystal Structure of the TTC7B/Hyccin Complex | | Descriptor: | Hyccin, Tetratricopeptide repeat protein 7B | | Authors: | Wu, X, Baskin, J.M, Reinisch, K.M, De Camilli, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane.
Nat.Cell Biol., 18, 2016
|
|
4RE9
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
6XJY
 
 | |
6XJQ
 
 | | Crystal structure of a self-alkylating ribozyme - alkylated form with biotinylated epoxide substrate | | Descriptor: | 2-{[(4R)-4-hydroxyhexyl]oxy}ethyl 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoate, Fab HAVx Heavy Chain, Fab HAVx Light Chain, ... | | Authors: | Koirala, D, Piccirilli, J.A. | | Deposit date: | 2020-06-24 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Structural basis for substrate binding and catalysis by a self-alkylating ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
6XJZ
 
 | |
